Abstract
Fertilization is an essential biological process in sexual reproduction and comprises a series of molecular interactions between the sperm and egg1,2. The fusion of the haploid spermatozoon and oocyte is the culminating event in mammalian fertilization, enabling the creation of a new, genetically distinct diploid organism3,4. The merger of two gametes is achieved through a two-step mechanism in which the sperm protein IZUMO1 on the equatorial segment of the acrosome-reacted sperm recognizes its receptor, JUNO, on the egg surface4,5,6. This recognition is followed by the fusion of the two plasma membranes. IZUMO1 and JUNO proteins are indispensable for fertilization, as constitutive knockdown of either protein results in mice that are healthy but infertile5,6. Despite their central importance in reproductive medicine, the molecular architectures of these proteins and the details of their functional roles in fertilization are not known. Here we present the crystal structures of human IZUMO1 and JUNO in unbound and bound conformations. The human IZUMO1 structure exhibits a distinct boomerang shape and provides structural insights into the IZUMO family of proteins7. Human IZUMO1 forms a high-affinity complex with JUNO and undergoes a major conformational change within its N-terminal domain upon binding to the egg-surface receptor. Our results provide insights into the molecular basis of sperm–egg recognition, cross-species fertilization, and the barrier to polyspermy, thereby promising benefits for the rational development of non-hormonal contraceptives and fertility treatments for humans and other mammals.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Wassarman, P. M., Jovine, L. & Litscher, E. S. A profile of fertilization in mammals. Nat. Cell Biol. 3, E59–E64 (2001)
Evans, J. P. Sperm-egg interaction. Annu. Rev. Physiol. 74, 477–502 (2012)
Sutovsky, P. Sperm-egg adhesion and fusion in mammals. Expert Rev. Mol. Med. 11, e11 (2009)
Klinovska, K., Sebkova, N. & Dvorakova-Hortova, K. Sperm-egg fusion: a molecular enigma of mammalian reproduction. Int. J. Mol. Sci. 15, 10652–10668 (2014)
Inoue, N., Ikawa, M., Isotani, A. & Okabe, M. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature 434, 234–238 (2005)
Bianchi, E., Doe, B., Goulding, D. & Wright, G. J. JUNO is the egg Izumo receptor and is essential for mammalian fertilization. Nature 508, 483–487 (2014)
Ellerman, D. A. et al. Izumo is part of a multiprotein family whose members form large complexes on mammalian sperm. Mol. Reprod. Dev. 76, 1188–1199 (2009)
Yanagimachi, R. Fertility of mammalian spermatozoa: its development and relativity. Zygote 2, 371–372 (1994)
Gupta, S. K. et al. Mammalian zona pellucida glycoproteins: structure and function during fertilization. Cell Tissue Res. 349, 665–678 (2012)
Kim, E. et al. Sperm penetration through cumulus mass and zona pellucida. Int. J. Dev. Biol. 52, 677–682 (2008)
Grayson, P. & Civetta, A. Positive selection and the evolution of izumo genes in mammals. Int. J. Evol. Biol. 2012, 958164 (2012)
Marquardt, T. & Helenius, A. Misfolding and aggregation of newly synthesized proteins in the endoplasmic reticulum. J. Cell Biol. 117, 505–513 (1992)
Halaby, D. M., Poupon, A. & Mornon, J. The immunoglobulin fold family: sequence analysis and 3D structure comparisons. Protein Eng. 12, 563–571 (1999)
Wibowo, A. S. et al. Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition. Proc. Natl Acad. Sci. USA 110, 15180–15188 (2013)
Chen, C. et al. Structural basis for molecular recognition of folic acid by folate receptors. Nature 500, 486–489 (2013)
Han, L. et al. Divergent evolution of vitamin B9 binding underlies JUNO-mediated adhesion of mammalian gametes. Curr. Biol. 26, R100–R101 (2016)
Inoue, N. et al. Molecular dissection of IZUMO1, a sperm protein essential for sperm-egg fusion. Development 140, 3221–3229 (2013)
Ohto, U. et al. Structure of IZUMO1–JUNO reveals sperm–oocyte recognition during mammalian fertilization. Nature http://dx.doi.org/10.1038/nature18596 (this issue)
Monné, M., Han, L., Schwend, T., Burendahl, S. & Jovine, L. Crystal structure of the ZP-N domain of ZP3 reveals the core fold of animal egg coats. Nature 456, 653–657 (2008)
Han, L. et al. Insights into egg coat assembly and egg-sperm interaction from the X-ray structure of full-length ZP3. Cell 143, 404–415 (2010)
Chen, E. H. & Olson, E. N. Unveiling the mechanisms of cell-cell fusion. Science 308, 369–373 (2005)
Wilson, I. A., Skehel, J. J. & Wiley, D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289, 366–373 (1981)
Pérez-Vargas, J. et al. Structural basis of eukaryotic cell-cell fusion. Cell 157, 407–419 (2014)
Subramanian, R. P. & Geraghty, R. J. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc. Natl Acad. Sci. USA 104, 2903–2908 (2007)
Sathiyamoorthy, K. et al. Assembly and architecture of the EBV B cell entry triggering complex. PLoS Pathog. 10, e1004309 (2014)
Inoue, N., Hagihara, Y., Wright, D., Suzuki, T. & Wada, I. Oocyte-triggered dimerization of sperm IZUMO1 promotes sperm–egg fusion in mice. Nat. Commun. 6, 8858 (2015)
Mohan, G. S., Li, W., Ye, L., Compans, R. W. & Yang, C. Antigenic subversion: a novel mechanism of host immune evasion by Ebola virus. PLoS Pathog. 8, e1003065 (2012)
Dolnik, O. et al. Ectodomain shedding of the glycoprotein GP of Ebola virus. EMBO J. 23, 2175–2184 (2004)
Whitmore, L. & Wallace, B. A. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32, W668–W673 (2004)
Sultana, A. & Lee, J. E. Measuring protein-protein and protein-nucleic acid interactions by biolayer interferometry. Curr. Prot. Protein Sci. 79, 19.25.1–19.26 (2015)
D’Arcy, A., Villard, F. & Marsh, M. An automated microseed matrix-screening method for protein crystallization. Acta Crystallogr. D 63, 550–554 (2007)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Potterton, E., Briggs, P., Turkenburg, M. & Dodson, E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D 59, 1131–1137 (2003)
Zwart, P. H., Grosse-Kunstleve, R. W., Lebedev, A. A., Murshudov, G. N. & Adams, P. D. Surprises and pitfalls arising from (pseudo)symmetry. Acta Crystallogr. D 64, 99–107 (2008)
Dauter, Z. Twinned crystals and anomalous phasing. Acta Crystallogr. D 59, 2004–2016 (2003)
Yeates, T. O. Simple statistics for intensity data from twinned specimens. Acta Crystallogr. A 44, 142–144 (1988)
Fisher, R. G. & Sweet, R. M. Treatment of diffraction data from crystals twinned by merohedry. Acta Crystallogr. A 36, 755–760 (1980)
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine . Acta Crystallogr. D 68, 352–367 (2012)
Terwilliger, T. C. et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D 64, 61–69 (2008)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot . Acta Crystallogr. D 66, 486–501 (2010)
Terwilliger, T. C. et al. Iterative-build OMIT maps: map improvement by iterative model building and refinement without model bias. Acta Crystallogr. D 64, 515–524 (2008)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993)
Förster, S., Apostol, L. & Bras, W. Scatter: software for the analysis of nano- and mesoscale small-angle scattering. J. Appl. Crystallogr. 43, 639–646 (2010)
Rambo, R. P. & Tainer, J. A. Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 (2013)
Svergun, D. I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 (1999)
Volkov, V. V. & Svergun, D. I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 (2003)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Marsh, J. J. et al. Structural insights into fibrinogen dynamics using amide hydrogen/deuterium exchange mass spectrometry. Biochemistry 52, 5491–5502 (2013)
Sheerin, D. J. et al. Inter- and intra-molecular interactions of Arabidopsis thaliana DELLA protein RGL1. Biochem. J. 435, 629–639 (2011)
Li, S. et al. Mechanism of intracellular cAMP sensor Epac2 activation: cAMP-induced conformational changes identified by amide hydrogen/deuterium exchange mass spectrometry (DXMS). J. Biol. Chem. 286, 17889–17897 (2011)
Zhang, Z. & Smith, D. L. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 2, 522–531 (1993)
Acknowledgements
This work was supported by a CIHR Operating Grant (MOP-115066), an NSERC Discovery Grant (RGPIN 435607-13), an Ontario Early Researcher Award (ER-13-09-116), and a Canada Research Chair to J.E.L. S.L. is supported by grants from NIH (1U19AI117905, R01 GM020501 and R01 AI101436). Support for stipends was provided by University of Toronto and Ontario Graduate Scholarships to H.A. and an NSERC USRA to A.T. We thank W. Houry, T. Moraes and C. Spring for access to circular dichroism, SEC–MALS and SPR systems, respectively. This work is based upon X-ray data collected at beamline 08ID-1 at the Canadian Light Source (CLS) and Structural Genomics Consortium (SGC), and SAXS data acquired at the Advanced Light Source (ALS) SIBYLS beamline 12.3.1. The CLS is supported by NSERC, National Research Council of Canada, CIHR, the Province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan. The ALS is a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the US Department of Energy (Office of Basic Energy Sciences) through the Integrated Diffraction Analysis Technologies program (DE-AC02-05CH11231), supported by the DOE Office of Biological and Environmental Research; additional support comes from NIH project MINOS (R01GM105404). We thank F. Azimi, J. Cook, A. Dong and N. Ly for technical support, and E. Ollmann Saphire, A. S. Rocca and G. Bikopoulos for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
H.A. designed the project, performed all the cloning, mutagenesis, expression, purification, biophysical characterization and crystallization experiments, collected synchrotron X-ray diffraction data, and determined the crystal structures and SAXS reconstructions. A.S. assisted with BLI and SPR experiments and provided crystallographic guidance for refinement and validation of the crystal structures. A.T. assisted with protein expression and purification. H.A. prepared the samples and S.L. performed and analysed the DXMS data. J.E.L supervised the research and assisted with BLI experiments. H.A. and J.E.L. analysed and discussed all results, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks K. Melcher, M. Okabe and other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Conservation of IZUMO1 residues.
a, Alignment of IZUMO1 protein sequences from various mammals. IZUMO1 sequences from Homo sapiens (human; GenBank: BAD91012.1), Macaca mulatta (rhesus macaque; GenBank: EHH30233.1), Gorilla gorilla (gorilla; Uniprot: G3QFY5), Pan paniscus (bonobo; NCBI: XP_003814124.1), Callithrix jacchus (marmoset; Uniprot: F7H859), Chlorocebus sabaeus (green monkey; Uniprot: A0A0D9S2Z4), Papio anubis (baboon; Uniprot: A0A0A0MU86), Nomascus leucogenys (gibbon; Uniprot: G1QXF7), Mus musculus (mouse; GenBank: BAD91011.1), Rattus norvegicus (rat; GenBank: BAD91013.1), Ictidomys tridecemlineatus (squirrel; Uniprot: I3N2L9), Cavia porcellus (guinea pig; Uniprot: H0UTJ7), Ochotona princeps (pika; NCBI: XP_004597241.1), Oryctolagus cuniculus (rabbit; Uniprot: G1TVX5), Felis catus (cat; NCBI: XP_006941089.1), Canis familiaris (dog, Uniprot: F6UM65), Ailuropoda melanoleuca (giant panda, Uniprot: G1M882), Equus caballus (horse; Uniprot: F6YE25), Bos taurus (cow; Uniprot: E1BDA8), Sus scrofa (pig; Uniprot: F1RIQ7), Capra hircus (goat; Uniprot: C6ZEA2), Ovis aries (sheep; Uniprot: W5PRD0), Sorex araneus (shrew; NCBI: XP_004619786.1), Pteropus vampyrus (megabat; NCBI: XP_011372928.1), Loxodonta africana (African elephant; NCBI: XP_003406572.1), and Dasypus novemcinctus (armadillo; NCBI: XP_004451154.1) are aligned. Red boxes indicate complete conservation of a given amino acid. N-linked glycosylation sequons (N-X-S/T) are indicated by red-coloured Y-shaped symbols. Secondary structural elements observed in the crystal structure of IZUMO1 are shown as arrows for β-strands and coils for α-helices. Residues that interact with JUNO are identified with asterisks, with those that form salt bridges and hydrogen bonds highlighted in blue and green boxes, respectively. Cysteine pairs involved in disulfide bond formation are numbered in red underneath each sequence. b, Footprint of JUNO on the molecular surface of IZUMO1. c, d, Representation of surface residue conservation, calculated using ConSurf and the alignment of all mammalian IZUMO1 (c) or primate-only IZUMO1 (d) sequences from Extended Data Fig. 1a. Degree of residue conservation is coloured in a gradient from high (burgundy) to low (cyan) variability.
Extended Data Figure 2 Conservation of JUNO residues.
a, Alignment of JUNO protein sequences from various mammals. JUNO/FOLR-δ sequences from H. sapiens (human; NCBI: NP_001186135.1), M. mulatta (rhesus macaque; NCBI: NP_001180734.1), G. gorilla (gorilla; NCBI: XP_004052029.1), P. paniscus (bonobo; NCBI: XP_003813838.1), C. jacchus (marmoset; NCBI: XP_009005477.1), C. sabaeus (green monkey; Uniprot: A0A0D9S1B0), P. anubis (baboon; NCBI: XP_009185381.1), N. leucogenys (gibbon; Uniprot: G1R639), M. musculus (mouse; NCBI: NP_075026.1), R. norvegicus (rat; NCBI: XP_001072998.2), I. tridecemlineatus (squirrel; NCBI: XP_005337246.1), C. porcellus (guinea pig; NCBI: XP_003468609.1), Cricetulus griseus (Chinese hamster; NCBI: XP_003506544.1) O. princeps (pika; NCBI: XP_012782378.1), O. cuniculus (rabbit; Uniprot: G1T5D7), F. catus (cat; NCBI: XP_011284828.1), C. familiaris (dog, Uniprot: E2RTK1), E. caballus (horse; NCBI: XP_001491306.1), S. scrofa (pig; Uniprot: F1STK4), C. hircus (goat; NCBI: XP_013824827.1), L. africana (African elephant; NCBI: XP_010593777.1), and D. novemcinctus (armadillo; NCBI: XP_004471965.1) are aligned. Red boxes indicate complete conservation of a given amino acid. N-linked glycosylation sequons (N-X-S/T) are indicated by red-coloured Y-shaped symbols. JUNO is anchored to the plasma membrane through a GPI anchor at Ser228 (shown as a green lollipop). Secondary structural elements observed in the crystal structure of JUNO are shown as arrows for β-strands and coils for α-helices. Residues that interact with IZUMO1 are identified with asterisks underneath the sequence, with those that form salt bridges and hydrogen bonds highlighted in blue and green boxes, respectively. Cysteine pairs involved in disulfide bond formation are numbered in red underneath each sequence. b, Footprint of IZUMO1 on the molecular surface of JUNO. c, d, Representation of surface residue conservation, calculated using ConSurf and the alignment of all mammalian JUNO (c) or primate-only JUNO sequences (d) from Extended Data Fig. 2a. Degree of residue conservation is coloured in a gradient from high (burgundy) to low (cyan) variability.
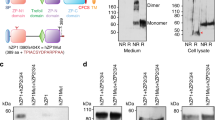
Extended Data Figure 3 Purification and characterization of IZUMO1 and JUNO.
a, Superdex-75 10/300 GL size-exclusion chromatograms of JUNO20–228, IZUMO122–254, and the IZUMO122–254–JUNO20–228 complex. Eluted peak positions of protein standards are marked with triangles and dashed lines. b, Coomassie-stained SDS–PAGE analysis of the purified IZUMO122–254, JUNO20–228 and IZUMO122–254–JUNO20–228 complex. For gel source data, see Supplementary Fig. 1c. c, Size-exclusion chromatography with inline multi-angle light scattering (SEC–MALS) profile of glycosylated human IZUMO122–268. The detector response unit (mV) and molecular mass (kDa) are plotted against the elution volume from a Superdex-200 Increase 10/300 GL size exclusion column. SEC-MALS reveals an apparent molecular mass of 34.8 kDa (dashed blue line), which corresponds to a monomeric species. d, Surface plasmon resonance (SPR) binding affinity and kinetic analysis of the human IZUMO122–254 and JUNO20–228 interaction. Human JUNO20–228 was amine-coupled to the SPR sensor chip. Kinetic parameters were derived from a Langmuir 1:1 binding model. e, Biolayer interferometry (BLI) kinetic analysis of the interaction between human IZUMO122–254 and JUNO20–228. Human JUNO20–228 was biotinylated and coupled to streptavidin-coated biosensors. Kinetic parameters were derived from a 1:1 binding model. The experimental curves are shown in colour superimposed with the fitted curves indicated as grey lines. f, A size distribution histogram from dynamic light scattering (DLS) measurements of IZUMO122–254, JUNO20–228 and IZUMO122–254–JUNO20–228 complex at 5 mg ml−1. IZUMO122–254, JUNO20–228 and IZUMO122–254–JUNO20–228 display hydrodynamic radii (RH) of ~3.0 nm, ~2.9 nm and ~3.9 nm, respectively. g, Circular dichroism (CD) wavelength scan of human IZUMO122–268 (blue) at 25 °C shows mixed secondary structural characteristics. The crystal structure of IZUMO122–268 aligns well with the secondary structural content calculated from the CD spectrum (35% α-helical, 24% β-strand and 41% random coil). A reconstructed CD wavelength scan (red) illustrates the agreement of the fit used in secondary structural content analysis. A CD thermal denaturation profile of human IZUMO122–268 at 222 nm is shown. The CD signal was normalized between 0 (folded) and 1 (unfolded), and plotted as a function of temperature. The Tm value indicates the midpoint of the melting transition.
Extended Data Figure 4 Structural comparison of JUNO and the folate receptor family of proteins.
a, Structural superimposition of JUNO20–228 with FOLR-α (PDB ID: 4LRH) and FOLR-β (PDB ID: 4KMZ). Experimentally bound folate (FOL), shown in white sticks, from the FOLR-α structure is positioned in the active site. b, Superimposition of residues in the folate-binding site of human FOLR-α and FOLR-β, and equivalent residues in human JUNO. Residue names shown in black are conserved among JUNO, FOLR-α and FOLR-β, and are numbered on the basis of the FOLR-α sequence. Inset boxes highlight the residue differences between JUNO, FOLR-α and FOLR-β. Key hydrogen bond interactions are shown as dashed black lines. Mutagenesis studies showed that replacement of D103 or D97 in FOLR-α or FOLR-β, respectively, which form strong interactions to the N1 and N2 nitrogen atoms of the pterin moiety, results in a decrease in affinity of more than one order of magnitude15. Six folate-binding residues observed in FOLR-α and FOLR-β (FOLR-α/FOLR-β: D103/D97, W124/W118, R125/R119, V129/F123, H157/H151, and K158/R152) are not conserved in JUNO. Four of these residues (FOLR-α/FOLR-β: D103/D97, W124/W118, R125/R119, and H157/H151) form key hydrogen bonds to anchor folate in the active site. In JUNO, the substituted residues are not able to maintain the extensive hydrogen bond network seen in FOLR-α and FOLR-β to folate. c, H. sapiens FOLR-α (Uniprot: P15328), FOLR-β (Uniprot: P14207), FOLR-γ (Uniprot: P41439) and FOLR-δ (Uniprot: A6ND01) are aligned. Red boxes indicate complete conservation of a given amino acid. N-linked glycosylation sequons (N-X-S/T) are indicated by red-coloured Y-shaped symbols. JUNO is anchored to the plasma membrane through a GPI anchor at Ser228 (shown as a green lollipop). Experimentally determined secondary structural elements are shown as arrows for β-strands and coils for α-helices. Key folate-binding residues, identified from the FOLR-α and FOLR-β crystal structures, are identified with an asterisk underneath the sequence. Key residue differences between JUNO, FOLR-α and FOLR-β folate binding sites are highlighted in a blue box.
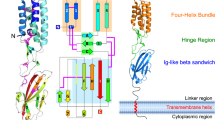
Extended Data Figure 5 IZUMO1–JUNO interface.
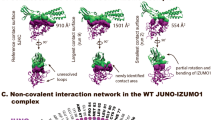
a, 2D schematic of the interactions between IZUMO122–254 and JUNO20–228. Residues from the IZUMO1 4HB, hinge, and Ig-like regions and from JUNO are coloured orange, green, blue and purple, respectively. Hydrogen-bond interactions are shown as dashed lines, and van der Waals forces are depicted as grey semi-circles. b, Footprints of JUNO on the surface of IZUMO1 and of IZUMO1 on the surface of JUNO. The molecular surfaces of IZUMO1 and JUNO are coloured white with residues forming interactions coloured as in a. No N-linked glycans on either IZUMO122–254 or JUNO20–228 are involved in binding. Formation of this interface results in a calculated free energy gain of −10.4 kcal mol−1.
Extended Data Figure 6 Hybrid structural analysis of human IZUMO1 and JUNO in a solution state.
a–c, Ab initio SAXS reconstruction, experimental scattering curves, normalized pair distance distribution function, P(r) and Kratky plot showing the degree of flexibility of IZUMO122–254 (a), JUNO20–228 (b), and the IZUMO122–254–JUNO20–228 complex (c). No concentration-dependent or radiation effects were observed in the SAXS data. The inset box in the experimental scattering data shows linearity in the Guinier plot at low q (qRg < 1.3). The IZUMO122–254, JUNO20–228 and IZUMO122–254–JUNO20–228 complex crystal structures were docked into the SAXS reconstructed molecular envelopes. The boomerang shape and upright conformation seen in the crystal structures of unbound and bound IZUMO122–254, respectively, were recapitulated by the SAXS reconstructions. d, Summary of the experimentally derived SAXS parameters for IZUMO122–254, JUNO20–228 and IZUMO122–254–JUNO20–228. The program SCATTER47 was used to calculate the radius of gyration (Rg) and maximum linear dimension (Dmax), and to perform Porod–Debye analysis to obtain the Porod volume and P coefficient. e, f, Comparative deuterium exchange mass spectrometry (DXMS) profile of unbound and bound IZUMO122–254 (e) and JUNO20–228 (f). The plots reveal the change in individual deuterium exchange for all observable residues. The coloured lines above the residue numbers correspond to the observed regions in the crystal structures.
Extended Data Figure 7 SAXS reconstruction of IZUMO1 and JUNO mutants.
a–d, Ab initio SAXS reconstruction, experimental scattering curves, normalized pair distance distribution function, P(r) and Kratky plot showing the degree of flexibility of wild-type (WT) IZUMO122–254–JUNO20–228(E45K) (a), IZUMO122–254(WT)–JUNO20–228(K163E) (b), IZUMO122–254(E71K)–JUNO20–228(WT) (c), and IZUMO122–254(R160E)–JUNO20–228(WT) (d) complexes. No concentration-dependent or radiation effects were observed. The inset box shows linearity in the Guinier plot at low q (qRg < 1.3). The IZUMO122–254(WT)–JUNO20–228(WT) complex crystal structure was docked into the SAXS reconstructed molecular envelopes. e, Summary of the experimentally derived SAXS parameters for the various IZUMO1–JUNO complexes. The program SCATTER47 was used to calculate the radius of gyration (Rg) and maximum linear dimension (Dmax), and to perform Porod–Debye analysis to obtain the Porod volume and P coefficient.
Extended Data Figure 8 Comparison of IZUMO1 with selected viral fusogens.
A common feature of many viral fusogens is the presence of a hydrophobic fusion peptide or fusion loop. a, Kyte and Doolittle hydropathy plots were calculated for IZUMO1, HIV-1 gp160, influenza A virus HA, Ebola virus glycoprotein (GP), Dengue virus type 2 E, and herpes simplex virus-1 gB to detect the presence of hydrophobic regions. Class I and class II viral fusion glycoproteins contain three clear hydrophobic regions corresponding to the signal peptide (grey), fusion peptide or loop (red) and the transmembrane anchor (blue). For class III viral glycoproteins, the presence of a signal peptide and transmembrane anchor are clear, but the hydrophobic fusion loop is formed by two discontinuous regions. This results in a lower hydropathy scale that is more difficult to detect. Two regions of hydrophobic residues cluster at the tip of the glycoprotein (shown in red) and are thought to be the internal fusion loop. In all class I, II and III viral fusion glycoproteins, clustering of aromatic and hydrophobic residues in a loop or helical region is a hallmark feature of fusion proteins. In contrast, IZUMO1 clearly does not have any hydrophobic regions or structural features similar to the viral fusogens that could insert into the egg membrane. b, Molecular surface representation of class I, II, and III viral glycoproteins and IZUMO1. The fusion peptide or loop is shown as red sticks and also coloured red on the glycoprotein surface. For the class I viral glycoproteins, the metastable prefusion trimer is shown, with the receptor binding and fusion subunits shown in blue and green, respectively. For the class II and class III viral glycoproteins, the postfusion trimer is shown with three hydrophobic fusion loops clustered at the tip of the molecule.
Extended Data Figure 9 Model of IZUMO1 and JUNO in sperm–egg fertilization.
During fertilization, mature sperm undergoes an acrosome reaction and penetrates through the egg zona pellucida to reach the perivitelline space. The acrosome reaction also causes relocalization of IZUMO1 to the sperm equatorial segment. a, IZUMO1 adopts a monomeric boomerang conformation on the surface of the sperm membrane. b, Upon binding to the JUNO egg receptor, IZUMO1 undergoes a conformational change. The 4HB region migrates towards the egg membrane. Moreover, the hinge region of IZUMO1 becomes more rigid and ‘locks’ the molecule into an upright position. The formation of the IZUMO1 and JUNO complex provides a direct physical link between the egg and sperm membranes. It is currently not clear whether IZUMO1 requires a post-JUNO binding event to trigger the fusion process, but at least three potential mechanisms are possible. c, The heterotypic assembly of IZUMO1 and JUNO, or a secondary conformational change in IZUMO1, may bring the egg and sperm membranes into close proximity for fusion to take place. d, Inoue et al. proposed that subsequent to IZUMO1–JUNO binding, a protein disulfide isomerase (PDI) catalyses a thio-disulfide exchange reaction that leads to structural conformation change and dimerization of IZUMO1 (ref. 26). The IZUMO1 dimer releases JUNO and contacts a yet-to-be-discovered oocyte receptor that facilitates membrane fusion. e, Alternatively, IZUMO1 may act as a scaffold to recruit other sperm or egg protein partners to form a multiprotein fusion complex in a manner similar to some viral fusogens. f, The merger of the egg and sperm membranes will require the apposition of the two bilayers to initiate initial mixing of the outer membrane leaflets and formation of a hemifusion stalk. The hemifused bilayers open to form the full fusion pore. g, Following fusion, JUNO is rapidly shed into extracellular vesicles from the fertilized oocyte. Within 30–40 min, JUNO is weakly or barely detectable on the membrane surface of zona-intact or anaphase II-stage zona-free fertilized oocytes, and undetectable at the pronuclear stage6. h, IZUMO1 binds JUNO tightly and rapidly (BLI: Kd = 59 ± 1 nM, ka = 1.15 × 105 M−1 s−1; SPR: 48 ± 4 nM, ka = 4.2 × 105 M−1 s−1), and once shed, JUNO is able to bind exposed IZUMO1 on incoming acrosomal-reacted sperm in the perivitelline space to act as a ‘sperm-sink’ to block polyspermy.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-9, Supplementary Table 1 and Supplementary References. (PDF 18568 kb)
Rights and permissions
About this article
Cite this article
Aydin, H., Sultana, A., Li, S. et al. Molecular architecture of the human sperm IZUMO1 and egg JUNO fertilization complex. Nature 534, 562–565 (2016). https://doi.org/10.1038/nature18595
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature18595
This article is cited by
-
Structural mechanism of mitochondrial membrane remodelling by human OPA1
Nature (2023)
-
Molecular dynamics of JUNO-IZUMO1 complexation suggests biologically relevant mechanisms in fertilization
Scientific Reports (2023)
-
SPACA6 ectodomain structure reveals a conserved superfamily of gamete fusion-associated proteins
Communications Biology (2022)
-
Live imaging-based assay for visualising species-specific interactions in gamete adhesion molecules
Scientific Reports (2022)
-
ASTL is mutated in female infertility
Human Genetics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.