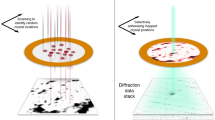
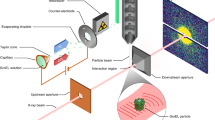
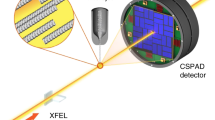
Abstract
The three-dimensional structures of macromolecules and their complexes are mainly elucidated by X-ray protein crystallography. A major limitation of this method is access to high-quality crystals, which is necessary to ensure X-ray diffraction extends to sufficiently large scattering angles and hence yields information of sufficiently high resolution with which to solve the crystal structure. The observation that crystals with reduced unit-cell volumes and tighter macromolecular packing often produce higher-resolution Bragg peaks1,2 suggests that crystallographic resolution for some macromolecules may be limited not by their heterogeneity, but by a deviation of strict positional ordering of the crystalline lattice. Such displacements of molecules from the ideal lattice give rise to a continuous diffraction pattern that is equal to the incoherent sum of diffraction from rigid individual molecular complexes aligned along several discrete crystallographic orientations and that, consequently, contains more information than Bragg peaks alone3. Although such continuous diffraction patterns have long been observed—and are of interest as a source of information about the dynamics of proteins4—they have not been used for structure determination. Here we show for crystals of the integral membrane protein complex photosystem II that lattice disorder increases the information content and the resolution of the diffraction pattern well beyond the 4.5-ångström limit of measurable Bragg peaks, which allows us to phase5 the pattern directly. Using the molecular envelope conventionally determined at 4.5 ångströms as a constraint, we obtain a static image of the photosystem II dimer at a resolution of 3.5 ångströms. This result shows that continuous diffraction can be used to overcome what have long been supposed to be the resolution limits of macromolecular crystallography, using a method that exploits commonly encountered imperfect crystals and enables model-free phasing6,7.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Heras, B. & Martin, J. L. Post-crystallization treatments for improving diffraction quality of protein crystals. Acta Crystallogr. D61, 1173–1180 (2005)
Umena, Y., Kawakami, K., Shen, J.-R. & Kamiya, N. Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60 (2011)
Sayre, D. Some implications of a theorem due to Shannon. Acta Crystallogr. 5, 843 (1952)
Wall, M. E., Adams, P. D., Fraser, J. S. & Sauter, N. K. Diffuse X-ray scattering to model protein motions. Structure 22, 182–184 (2014)
Fienup, J. R. Phase retrieval algorithms: a comparison. Appl. Opt. 21, 2758–2769 (1982)
Chapman, H. N. & Nugent, K. A. Coherent lensless X-ray imaging. Nature Photon . 4, 833–839 (2010)
Miao, J., Ishikawa, T., Robinson, I. K. & Murnane, M. M. Beyond crystallography: diffractive imaging using coherent x-ray light sources. Science 348, 530–535 (2015)
Guinier, A. X-Ray Diffraction in Crystals, Imperfect Crystals, and Amorphous Bodies Ch. 7 (Dover, 1994)
Rupp, B. Biomolecular Crystallography: Principles, Practice, and Application to Structural Biology Ch. 7, 10 (Garland Science, 2009)
Doucet, J. & Benoit, J. P. Molecular dynamics studied by analysis of the X-ray diffuse scattering from lysozyme crystals. Nature 325, 643–646 (1987)
Caspar, D. L. D., Clarage, J., Salunke, D. M. & Clarage, M. Liquid-like movements in crystalline insulin. Nature 332, 659–662 (1988)
Faure, P. et al. Correlated intramolecular motions and diffuse X-ray scattering in lysozyme. Nature Struct. Biol. 1, 124–128 (1994)
Pérez, J., Faure, P. & Benoit, J.-P. Molecular rigid-body displacements in a tetragonal lysozyme crystal confirmed by X-ray diffuse scattering. Acta Crystallogr. D52, 722–729 (1996)
Wall, M. E., Ealick, S. E. & Gruner, S. M. Three-dimensional diffuse X-ray scattering from crystals of Staphylococcal nuclease. Proc. Natl Acad. Sci. USA 94, 6180–6184 (1997)
Welberry, T. R., Heerdegen, A. P., Goldstone, D. C. & Taylor, I. A. Diffuse scattering resulting from macromolecular frustration. Acta Crystallogr. B67, 516–524 (2011)
Elser, V. & Millane, R. P. Reconstruction of an object from its symmetry-averaged diffraction pattern. Acta Crystallogr. A64, 273–279 (2008)
Bates, R. H. T. Fourier phase problems are uniquely solvable in more than one dimension. I: underlying theory. Optik 61, 247–262 (1982)
Fienup, J. R. Reconstruction of a complex-valued object from the modulus of its Fourier transform using a support constraint. J. Opt. Soc. Am. A 4, 118–123 (1987)
Elser, V. Phase retrieval by iterated projections. J. Opt. Soc. Am. A 20, 40–55 (2003)
Miao, J., Charalambous, P., Kirz, J. & Sayre, D. Extending the methodology of X-ray crystallography to allow imaging of micrometre-sized non-crystalline specimens. Nature 400, 342–344 (1999)
Robinson, I. K., Vartanyants, I. A., Williams, G. J., Pfeifer, M. A. & Pitney, J. A. Reconstruction of the shapes of gold nanocrystals using coherent X-ray diffraction. Phys. Rev. Lett. 87, 195505 (2001)
Chapman, H. N. et al. High-resolution ab initio three-dimensional X-ray diffraction microscopy. J. Opt. Soc. Am. A 23, 1179–1200 (2006)
Kupitz, C. et al. Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser. Nature 513, 261–265 (2014)
Chapman, H. N. et al. Femtosecond X-ray protein nanocrystallography. Nature 470, 73–77 (2011)
Boutet, S. et al. High-resolution protein structure determination by serial femtosecond crystallography. Science 337, 362–364 (2012)
White, T. A. et al. CrystFEL: a software suite for snapshot serial crystallography. J. Appl. Cryst. 45, 335–341 (2012)
Yefanov, O. et al. Mapping the continuous reciprocal space intensity distribution of X-ray serial crystallography. Phil. Trans. R. Soc. B 369, 20130333 (2014)
Shapiro, D. et al. Biological imaging by soft X-ray diffraction microscopy. Proc. Natl Acad. Sci. USA 102, 15343–15346 (2005)
Fischer, N. et al. Structure of the E. coli ribosome–EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM. Nature 520 , 567– 570 ( 2015).
Spence, J. C. H. & Doak, R. B. Single molecule diffraction. Phys. Rev. Lett. 92, 198102 (2004)
Liang, M. et al. The coherent X-ray imaging instrument at the Linac Coherent Light Source. J. Synchrotron Rad 22, 514–519 (2015)
Emma, P. et al. First lasing and operation of an ångstrom-wavelength free-electron laser. Nature Photon . 4, 641–647 (2010)
DePonte, D. P. et al. Gas dynamic virtual nozzle for generation of microscopic droplet streams. J. Phys. D 41, 195505 (2008)
Weierstall, U., Spence, J. C. H. & Doak, R. B. Injector for scattering measurements on fully solvated biospecies. Rev. Sci. Instrum. 83, 035108 (2012)
Neutze, R., Wouts, R., van der Spoel, D., Weckert, E. & Hajdu, J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature 406, 752–757 (2000)
Barty, A. et al. Self-terminating diffraction gates femtosecond X-ray nanocrystallography measurements. Nature Photon . 6, 35–40 (2012)
Hart, P. et al. The CSPAD megapixel x-ray camera at LCLS. In X-Ray Free-Electron Lasers: Beam Diagnostics, Beamline Instrumentation, and Application (eds Moeller, S. P. et al.) Proc. SPIE 8504, 85040C (SPIE, 2012)
Barty, A. et al. Cheetah: software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J. Appl. Cryst. 47, 1118–1131 (2014)
White, T. A. et al. Crystallographic data processing for free-electron laser sources. Acta Crystallogr. D69, 1231–1240 (2013)
Yefanov, O. et al. Accurate determination of segmented X-ray detector geometry. Opt. Express 23, 28459–28470 (2015)
Karplus, P. A. & Diederichs, K. Linking crystallographic model and data quality. Science 336, 1030–1033 (2012)
Cowley, J. M. Diffraction Physics 2nd edn, 150–151 (Elsevier, 1981)
Wilson, A. J. C. The probability distribution of X-ray intensities. Acta Crystallogr. 2, 318–321 (1949)
French, G. S. & Wilson, K. S. On the treatment of negative intensity observations. Acta Crystallogr. A34, 517–525 (1978)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D66, 213–221 (2010)
Urzhumtsev, A., Afonine, P. V., Lunin, V. Y., Terwilliger, T. C. & Adams, P. D. Metrics for comparison of crystallographic maps. Acta Crystallogr. D70, 2593–2606 (2014)
Acknowledgements
We acknowledge support of the Helmholtz Association through project-oriented funds and the Virtual Institute “Dynamic Pathways in Multidimensional Landscapes”; the DFG through the Gottfried Wilhelm Leibniz Program; the European Research Council under the European Union’s Seventh Framework Programme ERC Synergy Grant 609920 “AXSIS” and Marie Curie FP7-PEOPLE-2011-ITN Grant 317079 “Nanomem”; the BMBF through Project 05E13GU1; the Graduate College “GRK 1355” at the University of Hamburg, the International Max Planck Research School UFAST, the BioXFEL Science Technology Center (award 1231306); and the US National Institutes of Health (NIH), National Institute of General Medical Sciences grants R01 GM095583, U54 GM094599, and R01 GM097463. Parts of the sample injector used at the Linac Coherent Light Source (LCLS) for this research was funded by the NIH, P41GM103393, formerly P41RR001209. Use of the LCLS, SLAC National Accelerator Laboratory, is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract number DE-AC02-76SF00515.
Author information
Authors and Affiliations
Contributions
H.N.C. conceived the idea of molecular imaging using imperfect crystals, and developed it with K.A., A.B., O.M.Y., D.O., V.M., K.R.B. and L.G. K.A., A.B., O.M.Y., D.O., L.G. and V.M. carried out the diffractive imaging analysis. P.F. led the photosystem II project. Serial femtosecond crystallography measurements were led by P.F. and A.B. and performed with S.R.-C., J.C., C.K., D.O., M.M., A.S., K.D., D.J., G.N., U.W., Y.Z. and M.S. M.L., A.A., M.S.H., J.S.R., J.E.K. and S.Bo. set up and operated the CXI instrument. S.Ba., R.F., A.B. and M.M. performed on-site data evaluation at LCLS. S.Ba. and R.F. provided the initial search model and carried out model refinement on the image with D.O. Photosystem II was isolated by S.R.-C., J.C., C.E.C., A.S., C.K., A.S., J.-H.Y. and K.D., from which microcrystals were grown by S.R.-C., J.C., C.E.C., A.S. and C.K. Test experiments on crystals were performed by K.D., I.S., P.L.X. and M.M. H.N.C., A.B., K.A., O.M.Y., D.O., L.G., P.F., J.C.H.S., T.A.W., S.Ba. and S.Bo. wrote the manuscript with improvements from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Model refinement is improved at low resolution.
A plot of the metric Rfree as a function of resolution shell q, showing a marked improvement of the model refined against the 3.5-Å diffractive image. The blue curve shows Rfree prior to the inclusion of the continuous diffraction and the red curve shows Rfree afterwards. Here Rfree is calculated using only Bragg intensities (which were excluded from the refinement) for a resolution below 4.5 Å.
Extended Data Figure 2 Data quality and resolution of Bragg diffraction.
Plot of the reduced Pearson correlation coefficient, CC* (ref. 41), as an estimate of the consistency of the integrated Bragg intensities determined from 25,585 indexed patterns. A value of CC* = 0.5 is reached at q = 0.23 Å−1, or a resolution of 4.3 Å.
Extended Data Figure 3 Strongest continuous diffraction occurs with strongest Bragg diffraction.
a, Two-dimensional histogram of patterns, sorted by the integrated counts in the continuous component of the diffraction pattern (in a q range of 0.22–0.34 Å−1) and the integrated signal in all detected Bragg peaks, for all 25,585 indexed patterns. We chose the 2,848 patterns with the strongest continuous diffraction signal above 17 X-ray counts (purple line) in the q region to generate the 3D continuous pattern shown in Fig. 2c. The featureless background due to scattering from the solvent contributes 10 X-ray counts (blue line). b, The measurement from the liquid jet without crystals. c, d, Two representative patterns with speckle counts above the mean solvent background, but not above the threshold (c), and one of the 2,848 strongest patterns (d), as indicated by the positions of the yellow circles in a.
Extended Data Figure 4 Patterson function of a disordered lattice.
A distorted lattice (left, black circles, with ideal positions in grey), with vectors (red arrows) connecting all lattice points that have difference vectors an −ak = (1, 1). On the right, the arrows from the left panel are translated in two ways: upper right, the heads and tails are both displaced from their ideal positions; bottom right, the tails are lined up, resulting in the distribution of head positions forming a broader Gaussian. In the limit of a large crystal, the resultant distribution is the autocorrelation of the displacement distribution. This process can be repeated for all difference vectors, leading to equation (3).
Extended Data Figure 5 Continuous diffraction exhibits Wilson statistics.
Histogram of merged continuous intensities in a q range of 0.22–0.25 Å−1. Above a background level of around one photon per pixel per pulse, the logarithm of the histogram follows a linear trend with negative slope, characteristic of the exponential distribution predicted by Wilson statistics43.
Extended Data Figure 6 Central sections of the 3D full-pattern diffraction volume.
a–c, Bragg intensities in planes normal to the three orthogonal reciprocal-space axes, qz, qy and qx, respectively, which were arbitrarily chosen to be parallel to the c*, b* and a* axes of the PSII crystal. The 4.5-Å extent of the Bragg peaks is indicated by the red circle. d–f, Full-pattern diffraction intensities in central sections normal to the same three orthogonal axes as in a–c, respectively, obtained from that 2,848 strongest snapshot diffraction patterns. g–i, Continuous diffraction intensities calculated for a single PSII dimer using the model refined from the 4.5-Å Bragg data, for the same set of orthogonal planes as in a–c, respectively. The intensities were calculated from the incoherent sum of the squared modulus of the 3D molecular transform of a single (uncrystallized) PSII dimer in each of the four orientations of the 222 point group. All panels are plotted on the same scale, with the experimental data (d–f) extending to q = 0.33 Å−1 at the centre edge. The agreement between each of d–f with the corresponding panel g–i is further evidence that the rigid structural unit is the PSII dimer.
Extended Data Figure 7 Real-space orthogonal slices.
a–c, Slices of the observed structure factor (Fo) electron density map, plotted as a grey scale, of a single PSII dimer obtained from the 4.5-Å Bragg intensities following model refinement of that data. Each slice is one pixel thick (1.5 Å) and is normal to the z, y and x real-space axes, respectively (conjugate to qz, qy and qx). d–f, Slices through the 3D real-space support constraint used to for iterative phasing. The support was generated by blurring the 4.5-Å-resolution electron density map by 2.2 Å and then thresholding to achieve a binary mask. g–i, Slices through the 3.5-Å-resolution image obtained by iterative phasing of the continuous diffraction, using the support constraint illustrated in d–f and Fig. 3d.
Extended Data Figure 8 The continuous pattern is consistent with diffraction from a rigid object in crystallographic orientations.
a, When the continuous diffraction intensities are substituted for intensities averaged over all orientations (that is, constant on surfaces of constant q), iterative phasing using the support constraint of Fig. 3d fails, as indicated by this plot of the FSC as a function of resolution for solutions (green) obtained from two independent phasing trials. The blue curve is the same FSC shown in Fig. 3e for comparison. The iterations never converged, so a phase retrieval transfer function for the control could not be generated. b, Plot of the cross correlation (CC) between the measured diffraction and that calculated from the determined electron density of a dimer, symmetrized by the four crystallographic orientations.
Extended Data Figure 9 Electron density maps of regions of the PSII dimer.
a–f, Electron density maps based on maximum likelihood structure factors (2mFo-DFc) obtained using the Bragg diffraction (model shaded green, left), the Bragg and continuous diffraction (model shaded blue, middle) and computed from pseudo-crystal refinement (model shaded brown, right). The maps are rendered at various density levels relative to the standard deviation of the overall density. a, b, Non-haem iron coordinated by two His residues from D1 (chain A) and two from D2 (chain D), contoured at 1.5σ (a) and 4σ (b). Neighbouring Tyr and Lys residues are displayed as well. c, Part of an α-helix (chain T), showing that the side chains (for example, Arg and Phe) fit better in the electron density when applying our new method (maps contoured at 1.5σ). d, Helices from chains Y and Z (maps contoured at 1.25σ). The density map shows more details and better agreement to the model when applying the analysis using the Bragg and continuous diffraction. e, Detailed view of a section of chain Z (maps contoured at 1.25σ). Using only the Bragg diffraction, no electron density is visible at this level around the side chains of Trp, Lys and Arg. Again, the model fits better into the map when using the continuous diffraction. f, Two chlorophylls and part of the transmembrane helix of chain C (maps contoured at 1.5σ). g, Matrix of Pearson correlation coefficients of electron density maps obtained from the model presented in ref. 2 (PDB, 3WU2), the model refined from Bragg diffraction and that obtained from the continuous diffraction.
Rights and permissions
About this article
Cite this article
Ayyer, K., Yefanov, O., Oberthür, D. et al. Macromolecular diffractive imaging using imperfect crystals. Nature 530, 202–206 (2016). https://doi.org/10.1038/nature16949
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature16949
This article is cited by
-
Synchronous RNA conformational changes trigger ordered phase transitions in crystals
Nature Communications (2021)
-
A single-model quality assessment method for poor quality protein structure
BMC Bioinformatics (2020)
-
Diffuse X-ray scattering from correlated motions in a protein crystal
Nature Communications (2020)
-
Molecular movie of ultrafast coherent rotational dynamics of OCS
Nature Communications (2019)
-
Quantum imaging with incoherently scattered light from a free-electron laser
Nature Physics (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.