Abstract
The high prevalence of sickle haemoglobin in Africa shows that malaria has been a major force for human evolutionary selection, but surprisingly few other polymorphisms have been proven to confer resistance to malaria in large epidemiological studies1,2,3. To address this problem, we conducted a multi-centre genome-wide association study (GWAS) of life-threatening Plasmodium falciparum infection (severe malaria) in over 11,000 African children, with replication data in a further 14,000 individuals. Here we report a novel malaria resistance locus close to a cluster of genes encoding glycophorins that are receptors for erythrocyte invasion by P. falciparum. We identify a haplotype at this locus that provides 33% protection against severe malaria (odds ratio = 0.67, 95% confidence interval = 0.60–0.76, P value = 9.5 × 10−11) and is linked to polymorphisms that have previously been shown to have features of ancient balancing selection, on the basis of haplotype sharing between humans and chimpanzees4. Taken together with previous observations on the malaria-protective role of blood group O1,2,3,5, these data reveal that two of the strongest GWAS signals for severe malaria lie in or close to genes encoding the glycosylated surface coat of the erythrocyte cell membrane, both within regions of the genome where it appears that evolution has maintained diversity for millions of years. These findings provide new insights into the host–parasite interactions that are critical in determining the outcome of malaria infection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Data deposits
Genotype and phenotype data underlying this manuscript have been deposited in the European Genome-phenome Archive under accession number EGAS00001001311. Access to individual-level genotype data is available by application to an Independent Data Access Committee: see http://www.malariagen.net/data. For further details of data underlying this manuscript, see http://www.malariagen.net/resource/14.
References
Malaria Genomic Epidemiology Network. Reappraisal of known malaria resistance loci in a large multicenter study. Nature Genet. 46, 1197–1204 (2014)
Timmann, C. et al. Genome-wide association study indicates two novel resistance loci for severe malaria. Nature 489, 443–446 (2012)
Band, G. et al. Imputation-based meta-analysis of severe malaria in three African populations. PLoS Genet. 9, e1003509 (2013)
Leffler, E. M. et al. Multiple instances of ancient balancing selection shared between humans and chimpanzees. Science 339, 1578–1582 (2013)
Fry, A. E. et al. Common variation in the ABO glycosyltransferase is associated with susceptibility to severe Plasmodium falciparum malaria. Hum. Mol. Genet. 17, 567–576 (2008)
The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012)
Teo, Y. Y., Small, K. S. & Kwiatkowski, D. P. Methodological challenges of genome-wide association analysis in Africa. Nature Rev. Genet. 11, 149–160 (2010)
Manske, M. et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 487, 375–379 (2012)
Jallow, M. et al. Genome-wide and fine-resolution association analysis of malaria in West Africa. Nature Genet. 41, 657–665 (2009)
Blumenfeld, O. O. & Huang, C. H. Molecular genetics of the glycophorin gene family, the antigens for MNSs blood groups: multiple gene rearrangements and modulation of splice site usage result in extensive diversification. Hum. Mutat. 6, 199–209 (1995)
Sim, B. K., Chitnis, C. E., Wasniowska, K., Hadley, T. J. & Miller, L. H. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science 264, 1941–1944 (1994)
Mayer, D. C. et al. Glycophorin B is the erythrocyte receptor of Plasmodium falciparum erythrocyte-binding ligand, EBL-1. Proc. Natl Acad. Sci. USA 106, 5348–5352 (2009)
Baum, J., Ward, R. H. & Conway, D. J. Natural selection on the erythrocyte surface. Mol. Biol. Evol. 19, 223–229 (2002)
Ko, W. Y. et al. Effects of natural selection and gene conversion on the evolution of human glycophorins coding for MNS blood polymorphisms in malaria-endemic African populations. Am. J. Hum. Genet. 88, 741–754 (2011)
Tarazona-Santos, E. et al. Population genetics of GYPB and association study between GYPB*S/s polymorphism and susceptibility to P. falciparum infection in the Brazilian Amazon. PLoS ONE 6, e16123 (2011)
Wang, H. Y., Tang, H., Shen, C. K. & Wu, C. I. Rapidly evolving genes in human. I. The glycophorins and their possible role in evading malaria parasites. Mol. Biol. Evol. 20, 1795–1804 (2003)
Stranger, B. E. et al. Patterns of cis regulatory variation in diverse human populations. PLoS Genet. 8, e1002639 (2012)
Gurdasani, D. et al. The African Genome Variation Project shapes medical genetics in Africa. Nature 517, 327–332 (2015)
Lanzillotti, R. & Coetzer, T. L. The 10 kDa domain of human erythrocyte protein 4.1 binds the Plasmodium falciparum EBA-181 protein. Malar. J. 5, 100 (2006)
Ségurel, L. et al. The ABO blood group is a trans-species polymorphism in primates. Proc. Natl Acad. Sci. USA 109, 18493–18498 (2012)
Su, Z., Cardin, N., The Wellcome Trust Case Control Consortium, P. & Marchini, J. A Bayesian method for detecting and characterizing allelic heterogeneity and boosting signals in genome-wide association studies. Stat. Sci. 24, 430–450 (2009)
Lee, P. H., O’Dushlaine, C., Thomas, B. & Purcell, S. M. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics 28, 1797–1799 (2012)
Karlsson, E. K., Kwiatkowski, D. P. & Sabeti, P. C. Natural selection and infectious disease in human populations. Nature Rev. Genet. 15, 379–393 (2014)
Otto, T. D. et al. Genome sequencing of chimpanzee malaria parasites reveals possible pathways of adaptation to human hosts. Nature Commun. 5, 4754 (2014)
Liu, W. et al. Origin of the human malaria parasite Plasmodium falciparum in gorillas. Nature 467, 420–425 (2010)
Prugnolle, F. et al. A fresh look at the origin of Plasmodium falciparum, the most malignant malaria agent. PLoS Pathog. 7, e1001283 (2011)
Rich, S. M. et al. The origin of malignant malaria. Proc. Natl Acad. Sci. USA 106, 14902–14907 (2009)
Joy, D. A. et al. Early origin and recent expansion of Plasmodium falciparum. Science 300, 318–321 (2003)
Gilbert, S. C. et al. Association of malaria parasite population structure, HLA, and immunological antagonism. Science 279, 1173–1177 (1998)
Binks, R. H. et al. Population genetic analysis of the Plasmodium falciparum erythrocyte binding antigen-175 (eba-175) gene. Mol. Biochem. Parasitol. 114, 63–70 (2001)
World Health Organization Severe falciparum malaria. Trans. R. Soc. Trop. Med. Hyg. 94 (suppl. 1), S1–S90 (2000)
Teo, Y. Y. Genotype calling for the Illumina platform. Methods Mol. Biol. 850, 525–538 (2012)
Giannoulatou, E., Yau, C., Colella, S., Ragoussis, J. & Holmes, C. C. GenoSNP: a variational Bayes within-sample SNP genotyping algorithm that does not require a reference population. Bioinformatics 24, 2209–2214 (2008)
Kent, W. J. BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002)
Bellenguez, C. et al. A robust clustering algorithm for identifying problematic samples in genome-wide association studies. Bioinformatics 28, 134–135 (2012)
Astle, W. & Balding, D. J. Population structure and cryptic relatedness in genetic association studies. Stat. Sci. 24, 451–471 (2009)
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007)
R Development Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2015)
Ross, M. T. et al. The DNA sequence of the human X chromosome. Nature 434, 325–337 (2005)
Delaneau, O., Zagury, J. F. & Marchini, J. Improved whole-chromosome phasing for disease and population genetic studies. Nature Methods 10, 5–6 (2013)
Howie, B., Fuchsberger, C., Stephens, M., Marchini, J. & Abecasis, G. R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nature Genet. 44, 955–959 (2012)
Howie, B. N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009)
Wakefield, J. Bayes factors for genome-wide association studies: comparison with P-values. Genet. Epidemiol. 33, 79–86 (2009)
Carrel, L., Cottle, A. A., Goglin, K. C. & Willard, H. F. A first-generation X-inactivation profile of the human X chromosome. Proc. Natl Acad. Sci. USA 96, 14440–14444 (1999)
Pirinen, M., Donnelly, P. & Spencer, C. C. A. Efficient computation with a linear mixed model on large-scale data sets with applications to genetic studies. Ann. Appl. Stat. 7, 369–390 (2013)
GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015)
Welter, D. et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–D1006 (2014)
Karolchik, D. et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32, D493–D496 (2004)
Manjurano, A. et al. USP38, FREM3, SDC1, DDC, and LOC727982 gene polymorphisms and differential susceptibility to severe malaria in Tanzania. J. Infect. Dis. 212, 1129–1139 (2015)
The Wellcome Trust Case Control Consortium et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nature Genet. 44, 1294–1301 (2012)
Hillier, L. W. et al. Generation and annotation of the DNA sequences of human chromosomes 2 and 4. Nature 434, 724–731 (2005)
Zeller, T. et al. Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS One 5, e10693 (2010)
Acknowledgements
The Malaria Genomic Epidemiology Network Project is supported by the Wellcome Trust (WT077383/Z/05/Z) and the Bill & Melinda Gates Foundation through the Foundations of the National Institutes of Health (NIH; 566) as part of the Grand Challenges in Global Health Initiative. The Resource Centre for Genomic Epidemiology of Malaria is supported by the Wellcome Trust (090770/Z/09/Z). This research was supported by the Medical Research Council (MRC; G0600718; G0600230), the Wellcome Trust Biomedical ethics Enhancement Award (087285) and Strategic Award (096527). D.P.K. receives support from the MRC (G19/9). C.C.A.S. was supported by a Wellcome Trust Career Development Fellowship (097364/Z/11/Z). The Wellcome Trust also provides core awards to The Wellcome Trust Centre for Human Genetics (075491/Z/04; 090532/Z/09/Z) and the Wellcome Trust Sanger Institute (077012/Z/05/Z and 098051). The Mali MRTC–BMP group is supported by a contract (N01AI85346) and a cooperative agreement (U19AI065683) from the National Institute of Allergy and Infectious Diseases (NIAID) and by a grant (D43TW001589) from the Fogarty International Centre, NIH to University of Maryland and University of Bamako and the Mali-NIAID/NIH International Center for Excellence in Research at the University of Sciences, Techniques, and Technology of Bamako. E.A. received partial funding from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 242095–EVIMalaR and the Central African Network for Tuberculosis, HIV/AIDS and Malaria (CANTAM) funded by the European and Developing Countries Clinical Trials Partnership (EDCTP). T.N.W. is funded by Senior Fellowships from the Wellcome Trust (076934/Z/05/Z and 091758/Z/10/Z) and through the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 242095–EVIMalaR. The KEMRI-Wellcome Trust Programme is funded through core support from the Wellcome Trust. C.M.N. is supported through a strategic award to the KEMRI-Wellcome Trust Programme by the Wellcome Trust (084538). Tanzania/Kilimanjaro Christian Medical College Joint Malaria Programme, Moshi, Tanzania received funding from MRC grant number G9901439. We acknowledge the work of B. Poudiougou and A. Niangaly for their help in collecting clinical data and biological samples for the Bamako study. We thank L. Jostins and M. Pirinen for advice on statistical analyses.
Author information
Authors and Affiliations
Consortia
Contributions
Details of author contributions are provided in the author list.
Corresponding authors
Ethics declarations
Competing interests
The author declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Sample collections included in the study.
a, Study sites and ethics approving institutions. b, Phenotypic makeup of discovery and replication samples from each site. ‘Uncomplicated’ refers to case individuals who were not identified as cerebral malaria (CM) or severe malarial anaemia (SMA) cases. ‘Both’ refers to individuals who have both CM and SMA phenotypes. c, Overall sample counts and number of samples excluded by each quality control (QC) criterion. Asterisk denotes the number of samples removed after explicitly including samples with low heterozygosity in Gambia. Dagger: the Kenyan cohort included parents of a subset of case samples; these were not used in subsequent analyses d–f, Plots of average genome-wide heterozygosity and missingness with outliers coloured, as outputted by the ABERRANT algorithm.
Extended Data Figure 2 Genotyped SNP quality control for the three discovery cohorts.
a, b, Total numbers of pre- and post-quality control SNPs on the autosomes (a) and the X chromosome (b), and numbers of SNPs excluded by each quality control (QC) criterion. Diff, the test of difference in frequency between males and females; HWE, Hardy–Weinberg equilibrium; Plate, plate test of association. Details of quality control are given in Methods. c, Plot showing the –log10(P values) for the genotypic association test in the discovery data including the first five principal components as covariates. Grey dots show SNPs that are removed due to the quality control as defined in Methods. The total fraction of SNPs removed from each cohort is given at the top of the plot.
Extended Data Figure 3 Imputation performance.
a, b, Empirical distribution of concordance and accuracy (r2) between typed and re-imputed SNPs in the three discovery cohorts. Solid lines represent SNPs with frequency below 5% and dashed lines represent SNPs with frequency of at least 5%. c, Per-sample concordance and accuracy (type 0 r2) across the whole genome, as estimated by re-imputing genotyped SNPs. Values are averaged over imputation chunks. d, Average accuracy between genotype and re-imputed SNPs in each cohort, plotted against frequency, in 1% frequency bins.
Extended Data Figure 4 Detail of population structure.
a–c, Top ten principal components (PCs) in Gambia (a), Malawi (b) and Kenya (c). Where ethnicity was reported, points are coloured by ethnicity for ethnicities with at least 50 samples. d, Logistic regression P values and direction of effect for the top ten PCs on severe malaria status in each cohort. e, q–q plots for additive model association test P values in Gambia, Malawi, Kenya, and for fixed-effect meta-analysis. Dashed lines represent the 99% CI computed marginally at each variant. Circles and points represent points lying respectively outside and inside the 99% CI. f, Comparison of association test P values for logistic regression (SNPTEST, x-axis) and linear mixed model (MMM, y-axis) for Gambia, Malawi, Kenya, and for fixed-effect meta-analysis. Variants in tier 1 are coloured blue, with the lead marker at the FREM3/GYPE region coloured red.
Extended Data Figure 5 Detail of Bayesian analysis of discovery cohorts.
a, Visualization of slices through the combined prior on effect sizes in three cohorts for mode-of-inheritance-specific models. Top row: slices through the prior effect size on Kenya (x-axis) and Malawi (y-axis) for constant effect size in The Gambia (panels). Bottom row: slices through the prior effect size on Kenya (x-axis) and The Gambia (y-axis) for constant effect size in Malawi (panels). Red lines represent a factor of 10 in the prior density. b, Comparison of BFavg (x-axis) with the minimum fixed-effect meta-analysis P value minimized across additive, dominant, recessive or heterozygote modes of inheritance (y-axis). Values are plotted on log10 and −log10 scales. Colour indicates the heterogeneity model of the model with the highest posterior weight. c, Sensitivity of BFavg to changes in prior. Plots show BFavg ratio (y-axis) plotted against one-dimensional parameterisations of the prior (x-axis), for the 32 autosomal SNPs in tier 1. Solid lines represent variants with MAF < 5% averaged across populations, and dashed lines represent variants with MAF ≥ 5%. Black dots indicate the lead marker at the FREM3/GYPE locus. Colour indicates the effect size, mode of inheritance, or heterogeneity model for the model with highest posterior weight under the GWAS prior. Dashed grey vertical lines indicate the x-axis value corresponding to the prior used in the GWAS, and one-half and twice that value. Plots are parameterized by (1) the prior standard deviation of the small-effect model keeping the prior standard deviation of the large and small-effect models in the ratio 0.75:0.2; (2–5) the prior weight on additive, dominant, recessive or heterozygote modes of inheritance; (6–10) the prior weight on fixed, correlated, independent, fixed-structured and correlated-structured models. For each parameterization prior, weights on other models are kept in the same relative proportion. For further details see Supplementary Note 4.
Extended Data Figure 6 Strongest regions of association in the Bayesian analysis of the three discovery cohorts.
Plot on left shows the log10 model-averaged Bayes Factor (BFavg). Table shows the SNP with the highest BFavg in each region (lead SNP), gene(s) of interest in the region, the model with the highest posterior weight at the lead SNP and its BF. Models are described in Methods; sml, big: small- and large-effects models; fix, cor, ind, fix-str, cor-str: fixed, correlated, independent, fixed-structured and correlated-structured effect models; add, dom, rec, het: additive, dominant, recessive, heterozygote models; fs: fixed-between-sex model. Coloured points indicate the OR and the protective allele frequency in Gambia (red), Malawi (green) and Kenya (blue). The right-hand columns indicate regions containing shared chimpanzee–human haplotypes or coding SNPs4 (ABPs), blood group genes, or immunoglobulin superfamily genes.
Extended Data Figure 7 Evidence for association in the FREM3/GYPE, INPP4B and ARL14 regions.
a, Evidence for association at directly typed SNPs. b, Posterior probability that variants in the FREM3/GYPE region are causal, assuming a single variant in the region is causal50, based on the BFavg for typed and imputed variants. Dashed lines indicate the 95% and 99% credible sets. See Fig. 1 legend for further details. c, Details of SNPs encoding the common MNS blood groups. Coordinates and alleles are with respect to the NCBI b37 human reference sequence. d, Evidence for possible independence of effects at the FREM3 and INPP4B loci in Kenya by conditional analysis. y-Axis represents −log10(association P value) conditional on the imputed dosage at rs184895969. Points are coloured by linkage disequilibrium with the top SNP rs13103597. e, Forest plot showing sample size, estimated OR and 95% CI for the lead imputed SNP (rs149373719) in INPP4B under an additive model of association. f, Bar plot showing the posterior weight on different models of heterogeneity at rs149373719 under the prior used in the GWAS, assuming an additive model of association. g, Forest plot showing evidence in both discovery and replication samples in the Sequenom data at rs77389579 in INPP4B. See Fig. 2 legend for further details.
Extended Data Figure 8 Sequence homology, alignability and structural variation in the glycophorin region.
a, Co-occurrence of short segments of DNA (k-mers; top triangle: k = 100, bottom triangle: k = 25) in the human reference sequence Each point represents a k-mer that maps to the locations indicated by the x- and y-axis positions, either on the same strand (black points) or opposite strands (red points). Green vertical lines in this and subsequent panels delineate the region of high homology surrounding the three glycophorins. b, The location of the lead GWAS marker, ABPs, and protein-coding genes in the region. c, Alignability of the 100-mer at each position of the reference, up to two mismatches. Values are taken from the UCSC genome browser mappability track and averaged over 5 kb bins. d, IMPUTE info measure in Kenya for variants with frequency at least 5%, averaged over 5 kb bins. e–g, Coverage for samples from YRI and LWK in 1000 Genomes Project Phase I carrying esv2662558, carrying esv2668125, or not carrying either deletion, respectively. Coverage for each individual is normalized by the mean coverage for that individual across chromosome 1, and only computed at positions with alignability = 1 for all 100-mers overlapping the position, and for reads with mapping quality at least 20. Values are averaged over 5 kb (grey) and 10 kb (blue) bins. Three samples with apparently erroneous calls in the 1000 Genomes Phase I genotype release are coloured (NA18519, red; NA19185, yellow; NA19222, green) and assigned to the status indicated by their coverage profile. Panel g represents a random sample of 30 individuals not carrying the deletion in addition to the two with erroneous genotype calls. Coverage computation was performed using the BAM files available from the 1000 Genomes Project in October 2014, downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data. Four African samples in the Phase I release were not assessed because they are not included in this directory.
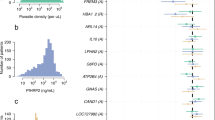
Extended Data Figure 9 Correlation between the genotypes at SNPs of interest within the GYPE/A/B locus and reported gene transcription levels in samples from the YRI and LWK HapMap cohorts17.
P values are for a trend test of association where more than one genotype class is present. Only assays targeting the glycophorins, and those with a P value below 0.05 are shown.
Extended Data Figure 10 Detail of enrichment analysis.
a, Red histogram: the empirical distribution of the log10 distance of observed tier 1 loci to the nearest ABP haplotype. Grey histogram, distribution of distances for 10,000 simulated tier 1 sets. b, The log10 distance of tier 1 (filled red circles) and tier 2 (empty circles) loci to the nearest ABP, plotted against their rank in BFavg order (stronger signals have lower rank). Loci are annotated with the nearest gene where a gene exists within the association region. Asterisks denote nearest genes that are also the nearest gene to an ABP shared haplotype. c, Empirical null distribution of the odds ratio for the enrichment of tier 1 loci in the set of genes closest to an ABP shared haplotype, based on 10,000 simulated SNP sets. The red asterisk and text indicate the odds ratio for the observed tier 1 loci. d, Distribution of the proportion of the genome which identifies a given gene as nearest, for genes in or not in the set annotated as nearest an ABP haplotype. Left, distribution of the length of the genome for which the given gene is unambiguously the closest gene. Middle, distribution of the number of SNPs in our study for which the given gene is the closest gene. Right, distribution of the number of SNPs in our study for which the given gene is the nearest gene within a recombination interval of 2.5 cM ± 25 kb around the SNP, as used to determine nearest genes to GWAS lead SNPs. e, Empirical P values for enrichment of ABP haplotypes and coding SNPs in tier 1 and tier 2 GWAS regions. Second column, P values for enrichment by gene overlap. Third to tenth column, P values for enrichment by proximity at different length scales. †Results for simulations using SNPs frequency-matched to GWAS tier 1 loci in 1% frequency bins. ††Results for simulations excluding the regions of ABO, HBB, ATP2B4, FREM3, INPP4B, and HHIP-AS1.
Supplementary information
Supplementary information
This file contains Supplementary Notes 1-4 and additional references. (PDF 183 kb)
Supplementary Table
This file contains a summary of statistics for 32 autosomal tier 1 loci for logistic regression and linear mixed model tests. (XLSX 51 kb)
Rights and permissions
About this article
Cite this article
Malaria Genomic Epidemiology Network. A novel locus of resistance to severe malaria in a region of ancient balancing selection. Nature 526, 253–257 (2015). https://doi.org/10.1038/nature15390
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature15390
This article is cited by
-
Genetic variations in human ATP2B4 gene alter Plasmodium falciparum in vitro growth in RBCs from Gambian adults
Malaria Journal (2023)
-
ATP2B4 regulatory genetic variants are associated with mild malaria
Malaria Journal (2023)
-
Genetics of cerebral malaria: pathogenesis, biomarkers and emerging therapeutic interventions
Cell & Bioscience (2022)
-
Risk score prediction model based on single nucleotide polymorphism for predicting malaria: a machine learning approach
BMC Bioinformatics (2022)
-
Leveraging Mann–Whitney U test on large-scale genetic variation data for analysing malaria genetic markers
Malaria Journal (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.