Abstract
G-protein-coupled receptors (GPCRs) signal primarily through G proteins or arrestins. Arrestin binding to GPCRs blocks G protein interaction and redirects signalling to numerous G-protein-independent pathways. Here we report the crystal structure of a constitutively active form of human rhodopsin bound to a pre-activated form of the mouse visual arrestin, determined by serial femtosecond X-ray laser crystallography. Together with extensive biochemical and mutagenesis data, the structure reveals an overall architecture of the rhodopsin–arrestin assembly in which rhodopsin uses distinct structural elements, including transmembrane helix 7 and helix 8, to recruit arrestin. Correspondingly, arrestin adopts the pre-activated conformation, with a ∼20° rotation between the amino and carboxy domains, which opens up a cleft in arrestin to accommodate a short helix formed by the second intracellular loop of rhodopsin. This structure provides a basis for understanding GPCR-mediated arrestin-biased signalling and demonstrates the power of X-ray lasers for advancing the frontiers of structural biology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Kim, Y. J. et al. Crystal structure of pre-activated arrestin p44. Nature 497, 142–146 (2013)
Shukla, A. K. et al. Structure of active β-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide. Nature 497, 137–141 (2013)
Pitcher, J. A., Freedman, N. J. & Lefkowitz, R. J. G protein-coupled receptor kinases. Annu. Rev. Biochem. 67, 653–692 (1998)
Wilden, U., Hall, S. W. & Kuhn, H. Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48-kDa protein of rod outer segments. Proc. Natl Acad. Sci. USA 83, 1174–1178 (1986)
Reiter, E., Ahn, S., Shukla, A. K. & Lefkowitz, R. J. Molecular mechanism of beta-arrestin-biased agonism at seven-transmembrane receptors. Annu. Rev. Pharmacol. Toxicol. 52, 179–197 (2012)
Kenakin, T. P. Biased signalling and allosteric machines: new vistas and challenges for drug discovery. Br. J. Pharmacol. 165, 1659–1669 (2012)
Palczewski, K. et al. Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289, 739–745 (2000)
Rasmussen, S. G. et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011)
Cherezov, V. et al. High-resolution crystal structure of an engineered human β2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 (2007)
Katritch, V., Cherezov, V. & Stevens, R. C. Structure-function of the G protein-coupled receptor superfamily. Annu. Rev. Pharmacol. Toxicol. 53, 531–556 (2013)
Standfuss, J. et al. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature 471, 656–660 (2011)
Xu, F. et al. Structure of an agonist-bound human A2A adenosine receptor. Science 332, 322–327 (2011)
Wang, C. et al. Structural basis for molecular recognition at serotonin receptors. Science 340, 610–614 (2013)
Wacker, D. et al. Structural features for functional selectivity at serotonin receptors. Science 340, 615–619 (2013)
Liu, J. J., Horst, R., Katritch, V., Stevens, R. C. & Wuthrich, K. Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science 335, 1106–1110 (2012)
Zhou, X. E., Melcher, K. & Xu, H. E. Structure and activation of rhodopsin. Acta Pharmacol. Sin. 33, 291–299 (2012)
Gurevich, V. V., Hanson, S. M., Song, X. F., Vishnivetskiy, S. A. & Gurevich, E. V. The functional cycle of visual arrestins in photoreceptor cells. Prog. Retin. Eye Res. 30, 405–430 (2011)
Smith, S. O. Insights into the activation mechanism of the visual receptor rhodopsin. Biochem. Soc. Trans. 40, 389–393 (2012)
Han, M., Smith, S. O. & Sakmar, T. P. Constitutive activation of opsin by mutation of methionine 257 on transmembrane helix 6. Biochemistry 37, 8253–8261 (1998)
Ballesteros, J. A. & Weinstein, H. Integrated methods for the construction of three dimensional models and computational probing of structure-function relations in G-protein coupled receptors. Methods in Neurosciences 25, 366–428 (1995)
Park, J. H., Scheerer, P., Hofmann, K. P., Choe, H. W. & Ernst, O. P. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 454, 183–187 (2008)
Scheerer, P. et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature 455, 497–502 (2008)
Choe, H. W. et al. Crystal structure of metarhodopsin II. Nature 471, 651–655 (2011)
Hirsch, J. A., Schubert, C., Gurevich, V. V. & Sigler, P. B. The 2.8 Å crystal structure of visual arrestin: a model for arrestin's regulation. Cell 97, 257–269 (1999)
Granzin, J. et al. X-ray crystal structure of arrestin from bovine rod outer segments. Nature 391, 918–921 (1998)
Shukla, A. K. et al. Visualization of arrestin recruitment by a G-protein-coupled receptor. Nature 512, 218–222 (2014)
Standfuss, J., Zaitseva, E., Mahalingam, M. & Vogel, R. Structural impact of the E113Q counterion mutation on the activation and deactivation pathways of the G protein-coupled receptor rhodopsin. J. Mol. Biol. 380, 145–157 (2008)
Xie, G., Gross, A. K. & Oprian, D. D. An opsin mutant with increased thermal stability. Biochemistry 42, 1995–2001 (2003)
Standfuss, J. et al. Crystal structure of a thermally stable rhodopsin mutant. J. Mol. Biol. 372, 1179–1188 (2007)
Martin, E. L., Rens-Domiano, S., Schatz, P. J. & Hamm, H. E. Potent peptide analogues of a G protein receptor-binding region obtained with a combinatorial library. J. Biol. Chem. 271, 361–366 (1996)
Zhuang, T. et al. Involvement of distinct arrestin-1 elements in binding to different functional forms of rhodopsin. Proc. Natl Acad. Sci. USA 110, 942–947 (2013)
Bayburt, T. H. et al. Monomeric rhodopsin is sufficient for normal rhodopsin kinase (GRK1) phosphorylation and arrestin-1 binding. J. Biol. Chem. 286, 1420–1428 (2011)
Hanson, S. M. et al. Each rhodopsin molecule binds its own arrestin. Proc. Natl Acad. Sci. USA 104, 3125–3128 (2007)
Boutet, S. et al. High-resolution protein structure determination by serial femtosecond crystallography. Science 337, 362–364 (2012)
Weierstall, U. et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nature Commun. 5, 3309 (2014)
Liu, W. et al. Serial femtosecond crystallography of G protein-coupled receptors. Science 342, 1521–1524 (2013)
Barty, A. et al. software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J. Appl. Crystallogr. 47, 1118–1131 (2014)
White, T. A. et al. CrystFEL: a software suite for snapshot serial crystallography. J. Appl. Crystallogr. 45, 335–341 (2012)
Deupi, X. et al. Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc. Natl Acad. Sci. USA 109, 119–124 (2012)
Altenbach, C., Kusnetzow, A. K., Ernst, O. P., Hofmann, K. P. & Hubbell, W. L. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc. Natl Acad. Sci. USA 105, 7439–7444 (2008)
Kim, M. et al. Conformation of receptor-bound visual arrestin. Proc. Natl Acad. Sci. USA 109, 18407–18412 (2012)
Kirchberg, K. et al. Conformational dynamics of helix 8 in the GPCR rhodopsin controls arrestin activation in the desensitization process. Proc. Natl Acad. Sci. USA 108, 18690–18695 (2011)
Ostermaier, M. K., Peterhans, C., Jaussi, R., Deupi, X. & Standfuss, J. Functional map of arrestin-1 at single amino acid resolution. Proc. Natl Acad. Sci. USA 111, 1825–1830 (2014)
Sommer, M. E., Hofmann, K. P. & Heck, M. Distinct loops in arrestin differentially regulate ligand binding within the GPCR opsin. Nature Commun. 3, 995 (2012)
West, G. M. et al. Protein conformation ensembles monitored by HDX reveal a structural rationale for abscisic acid signaling protein affinities and activities. Structure 21, 229–235 (2013)
Ohguro, H., Palczewski, K., Walsh, K. A. & Johnson, R. S. Topographic study of arrestin using differential chemical modifications and hydrogen-deuterium exchange. Protein Sci. 3, 2428–2434 (1994)
Barnea, G. et al. The genetic design of signaling cascades to record receptor activation. Proc. Natl Acad. Sci. USA 105, 64–69 (2008)
Gurevich, V. V. & Benovic, J. L. Visual arrestin interaction with rhodopsin. Sequential multisite binding ensures strict selectivity toward light-activated phosphorylated rhodopsin. J. Biol. Chem. 268, 11628–11638 (1993)
Fotiadis, D. et al. Atomic-force microscopy: rhodopsin dimers in native disc membranes. Nature 421, 127–128 (2003)
Zhang, H. et al. Structure of the angiotensin receptor revealed by serial femtosecond crystallography. Cell 161, 833–844 (2015)
Beckett, D., Kovaleva, E. & Schatz, P. J. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8, 921–929 (1999)
Mossessova, E. & Lima, C. D. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876 (2000)
Alexandrov, A. I., Mileni, M., Chien, E. Y. T., Hanson, M. A. & Stevens, R. C. Microscale fluorescent thermal stability assay for membrane proteins. Structure 16, 351–359 (2008)
Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nature Protocols 4, 706–731 (2009)
Chen, A. H., Hummel, B., Qiu, H. & Caffrey, M. A simple mechanical mixer for small viscous lipid-containing samples. Chem. Phys. Lipids 95, 11–21 (1998)
Xu, F., Liu, W., Hanson, M. A., Stevens, R. C. & Cherezov, V. Development of an automated high throughput LCP-FRAP assay to guide membrane protein crystallization in lipid mesophases. Cryst. Growth Des. 11, 1193–1201 (2011)
Liu, W., Ishchenko, A. & Cherezov, V. Preparation of microcrystals in lipidic cubic phase for serial femtosecond crystallography. Nature Protocols 9, 2123–2134 (2014)
Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D 66, 133–144 (2010)
Boutet, S. & Williams, G. J. The Coherent X-ray Imaging (CXI) instrument at the Linac Coherent Light Source (LCLS). New J. Phys. 12, 035024 (2010)
Siewert, F. et al. Ultra-precise characterization of LCLS hard X-ray focusing mirrors by high resolution slope measuring deflectometry. Opt. Express 20, 4525–4536 (2012)
White, T. A. et al. Crystallographic data processing for free-electron laser sources. Acta Crystallogr. D 69, 1231–1240 (2013)
Battye, T. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D 67, 271–281 (2011)
Duisenberg, A. J. M. Indexing in single-crystal diffractometry with an obstinate list of reflections. J. Appl. Crystallogr. 25, 92–96 (1992)
Kirian, R. A. et al. Structure-factor analysis of femtosecond microdiffraction patterns from protein nanocrystals. Acta Crystallogr. A 67, 131–140 (2011)
Karplus, P. A. & Diederichs, K. Linking crystallographic model and data quality. Science 336, 1030–1033 (2012)
Lebedev, A. A. & Isupov, M. N. Space-group and origin ambiguity in macromolecular structures with pseudo-symmetry and its treatment with the program Zanuda. Acta Crystallogr. D 70, 2430–2443 (2014)
Padilla, J. E. & Yeates, T. O. A statistic for local intensity differences: robustness to anisotropy and pseudo-centering and utility for detecting twinning. Acta Crystallogr. D 59, 1124–1130 (2003)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Tickle, I. J. Statistical quality indicators for electron-density maps. Acta Crystallogr. D 68, 454–467 (2012)
DeLano, W. L. & Lam, J. W. PyMOL: a communications tool for computational models. Abstr. Pap. Am. Chem. Soc. 230, U1371–U1372 (2005)
Moeller, A., Kirchdoerfer, R. N., Potter, C. S., Carragher, B. & Wilson, I. A. Organization of the influenza virus replication machinery. Science 338, 1631–1634 (2012)
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005)
Lander, G. C. et al. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009)
Voss, N. R., Yoshioka, C. K., Radermacher, M., Potter, C. S. & Carragher, B. DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 (2009)
Scheres, S. H. W., Nunez-Ramirez, R., Sorzano, C. O. S., Carazo, J. M. & Marabini, R. Image processing for electron microscopy single-particle analysis using XMIPP. Nature Protocols 3, 977–990 (2008)
Goswami, D. et al. Time window expansion for HDX analysis of an intrinsically disordered protein. J. Am. Soc. Mass Spectrom. 24, 1584–1592 (2013)
Pascal, B. D. et al. HDX Workbench: software for the analysis of H/D exchange MS data. J. Am. Soc. Mass Spectrom. 23, 1512–1521 (2012)
Klein-Seetharaman, J. et al. Single-cysteine substitution mutants at amino acid positions 55-75, the sequence connecting the cytoplasmic ends of helices I and II in rhodopsin: reactivity of the sulfhydryl groups and their derivatives identifies a tertiary structure that changes upon light-activation. Biochemistry 38, 7938–7944 (1999)
Li, Z., Michael, I. P., Zhou, D., Nagy, A. & Rini, J. M. Simple piggyBac transposon-based mammalian cell expression system for inducible protein production. Proc. Natl Acad. Sci. USA 110, 5004–5009 (2013)
Caro, L. N. et al. Rapid and facile recombinant expression of bovine rhodopsin in HEK293S GnTI(-) cells using a PiggyBac inducible system. Methods Enzymol. 556, 307–330 (2015)
Reeves, P. J., Callewaert, N., Contreras, R. & Khorana, H. G. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl Acad. Sci. USA 99, 13419–13424 (2002)
Wang, W. et al. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA 105, 9290–9295 (2008)
Bayburt, T. H., Leitz, A. J., Xie, G., Oprian, D. D. & Sligar, S. G. Transducin activation by nanoscale lipid bilayers containing one and two rhodopsins. J. Biol. Chem. 282, 14875–14881 (2007)
Hanson, S. M. et al. Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. Proc. Natl Acad. Sci. USA 103, 4900–4905 (2006)
Hanson, S. M. et al. Structure and function of the visual arrestin oligomer. EMBO J. 26, 1726–1736 (2007)
Fleissner, M. R., Cascio, D. & Hubbell, W. L. Structural origin of weakly ordered nitroxide motion in spin-labeled proteins. Protein Sci. 18, 893–908 (2009)
Lietzow, M. A. & Hubbell, W. L. Motion of spin label side chains in cellular retinol-binding protein: correlation with structure and nearest-neighbor interactions in an antiparallel beta-sheet. Biochemistry 43, 3137–3151 (2004)
Qiu, H. & Caffrey, M. The phase diagram of the monoolein/water system: metastability and equilibrium aspects. Biomaterials 21, 223–234 (2000)
Misquitta, Y. et al. Rational design of lipid for membrane protein crystallization. J. Struct. Biol. 148, 169–175 (2004)
Misquitta, L. V. et al. Membrane protein crystallization in lipidic mesophases with tailored bilayers. Structure 12, 2113–2124 (2004)
ICM. Manual v. 3.8 (MolSoft LLC, 2014)
Coin, I. et al. Genetically encoded chemical probes in cells reveal the binding path of urocortin-I to CRF class B GPCR. Cell 155, 1258–1269 (2013)
Arnautova, Y. A., Abagyan, R. A. & Totrov, M. Development of a new physics-based internal coordinate mechanics force field and its application to protein loop modeling. Proteins 79, 477–498 (2011)
Acknowledgements
Portions of this research were carried out at the Linac Coherent Light Source (LCLS) at the SLAC National Accelerator Laboratory. Use of the LCLS at the SLAC National Accelerator Laboratory is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract no. DE-AC02-76SF00515. Parts of the sample injector used at LCLS for this research was funded by the National Institutes of Health, P41GM103393, formerly P41RR001209. We thank staff members of the Life Science Collaborative Access Team (ID-21) of the Advanced Photon Source (APS) for assistance in data collection at the beam lines of sector 21, which is in part funded by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817), and the General Medicine Collaborative Access Team for assistance in data collection at the beam lines of sector 23 (ID-23), funded in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). Use of APS was supported by the Office of Science of the US Department of Energy, under contract no. DE-AC02-06CH11357. This work was supported in part by the Jay and Betty Van Andel Foundation, Ministry of Science and Technology (China) grants 2012ZX09301001 and 2012CB910403, 2013CB910600, XDB08020303, 2013ZX09507001, Amway (China), National Institute of Health grants, DK071662 (H.E.X.); GM073197 and GM103310 (C.S.P. and B.C.); GM102545 and GM104212 (K.M.); EY011500 and GM077561 (V.V.G.), EY005216 and P30 EY000331 (W.L.H.), the National Institutes of Health Common Fund in Structural Biology grants P50 GM073197 (V.C. and R.C.S.), P50 GM073210 (M.C.), and GM095583 (P.F.); National Institute of General Medical Sciences PSI: Biology grants U54 GM094618 (V.C., V.K., and R.C.S.), GM108635 (V.C.), U54 GM094599 (P.F.), GM097463 (J.S.), and U54 GM094586 (JCSG); NSF Science and Technology Center award 1231306 (J.C.H.S., P.F. and U.W.); Swiss National Science Foundation grant 31003A_141235 (J.S.); the Canada Excellence Research Chair program and the Anne & Max Tanenbaum Chair in Neuroscience at the University of Toronto (O.P.E.); and Science Foundation Ireland, grant 12/IA/1255 (M.C.). Parts of this work were also supported by the Helmholtz Gemeinschaft, the DFG Cluster of Excellence Center for Ultrafast Imaging, and the BMBF project FKZ 05K12CH1 (H.N.C., A.B., C.G., O.Y., T.W.); the Irene and Eric Simon Brain Research Foundation (R.L.). We thank A. Brunger and O. Zeldin for analysing the XFEL data and for advising on refinement; B. Weis for advice on twin refinement and structure validation; J. Rini for advice on the piggyBac expression system; A. Lebedev for his advice regarding the Zanuda program and the choice of the space group; and A. Walker for final editing of the manuscript. C.G. kindly thanks the PIER Helmholtz-Graduate School and the Helmholtz Association for financial support. We also thank the TianHe research and development team of National University of Defense Technology (NUDT) for computational resources.
Author information
Authors and Affiliations
Contributions
Y.K. initiated the project, developed the expression and purification methods for rhodopsin–arrestin complex, and bulk-purified expression constructs and proteins used in LCP crystallization for the SFX method; X.E.Z. collected the synchrotron data, helped with the SFX data collection, processed the data, and solved the structures; X. Gao expressed and purified rhodopsin–arrestin complexes, characterized their binding and thermal stability, discovered the initial crystallization conditions with 9.7 MAG (1-(9Z-hexadecenoyl)-rac-glycerol), prepared most crystals for synchrotron data collection, prepared all crystals for the final data collection by SFX, helped with SFX data collection, and established the initial cross-linking method for the rhodopsin–arrestin complex; Y.H. designed and performed Tango assays and disulfide bond cross-linking experiments; C.Z. developed the mammalian expression methods; P.W.d.W. helped with XFEL data processing and performed computational experiments; J.K., M.H.E.T., K.M.S.-P., K.P., J.M., Y.J., X.Z., and X. Gu performed cell culture, mutagenesis, protein purification, rhodopsin–arrestin binding experiments; W.L. and A.I. grew crystals and collected synchrotron data at APS and SFX data at LCLS, G.W.H. and Q.X. determined and validated the structure. Z.Z. and V.K. constructed the full model, the phosphorylated rhodopsin–arrestin model, and helped writing the paper; D.W., S.L., D.J., C.K., Sh.B., and N.A.Z. helped with XFEL data collection and initial data analysis; Sé.B., M.M., and G.J.W. set up the XFEL experiment, performed the data collection, and commented on the paper. A.B., T.A.W., C.G., O.Y., and H.N.C. helped with XFEL data collection and data analysis, processed the data and helped with structure validation. G.M. W., B.D.P., and P.R.G. performed HDX experiments and helped with manuscript writing. J.L. helped initiate this collaborative project and with writing the paper. M.W. collected the 7.7 Å dataset at the Swiss Light Source. A.M., C.S.P., and B.C. were responsible for electron microscopy images of rhodopsin–arrestin complexes. M.T. and Y.Z. performed mass spectrometry experiments to validate the protein contents in the crystals; D.L., N. H., and M.C. provided the 9.7 MAG phase diagram and helped with SFX data collection and with writing the paper. J.S. provided a computational model of the rhodopsin–arrestin complex and helped with discussion and writing; K.D., H.L., and Y.D. helped with data analysis and twinning problems; R.J.L. constructed single-Cys arrestin-1 mutants for DEER and tested their binding to rhodopsin; S.A.V. expressed these mutants in Escherichia coli and purified them; V.V.G. provided arrestin genes, designed single-Cys arrestin-1 mutants for DEER, and helped analysing the data and writing the paper. H.Y. and H.J. performed computational modelling, figure preparation, and helped with writing the paper; J.C.H.S. and U.W. designed the LCP injector and helped with data collection. Sh.B., S.R.-C., C.E.C., J.C., C.K., I.G., P.F., and R.F. helped with data collection, on-site crystal characterization as well as data analysis, and validation of the structure. L.N.C. and O.P.E. generated the Y74C/C140S/C316S stable cell line, characterized and provided the rhodopsin mutant sample for DEER measurements. N.V.E. and W.L.H. incorporated rhodopsin into nanodiscs, spin-labelled rhodopsin and arrestin, performed DEER experiments and helped with manuscript writing. R.C.S. supervised crystal growth, data collection, structure solution and validation, and helped with manuscript writing. V.C. was the Principal Investigator of the LCLS data collection, supervised crystal growth, data collection at APS and LCLS, structure solution and validation, and helped with manuscript writing; K.M. supervised research, analysed data, and helped with writing the paper. H.E.X. conceived the project, designed the research, performed synchrotron and LCLS data collection and structure solution, and wrote the paper with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
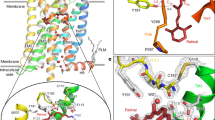
Extended Data Figure 1 Constitutively active rhodopsin interacts with arrestin and GαCT-HA.
a, SDS–PAGE of N-terminally MBP-tagged wild-type and mutant rhodopsin. b, Non-cropped versions of the pull-down assay gels shown in Fig. 1b. The interactions between mouse wild-type arrestin and human wild-type or E1133.28Q rhodopsin are very weak. In contrast, the interaction between constitutively active rhodopsin (E1133.28Q/M2576.40Y) and pre-activated L374A/V375A/F376A arrestin (3A arrestin) is strong and is further increased in the presence of 10 μM all-trans-retinal. Input: 5% of the binding reaction. Bottom panels show the rhodopsin loading controls. c, Schematic representation of the AlphaScreen assay. d, AlphaScreen binding assay between E1133.28Q/M2576.40Y rhodopsin and GαCT-HA (TGGRVLEDLKSCGLF) in the presence and absence of 5 µM all-trans-retinal. The two left columns show the controls with ‘peptide only’ and ‘rhodopsin only’. (n = 3, error bars, s.d.). e, Determination of the affinity of the interaction between rhodopsin E1133.28Q/M2576.40Y and GαCT-HA by homologous competition. His6–MBP–rhodopsin mutant protein was immobilized on Ni-acceptor beads and biotinylated GαCT-HA on streptavidin donor beads. Binding between rhodopsin and arrestin brings donor and acceptor beads into close proximity, resulting in the indicated binding signal. Non-biotinylated GαCT-HA competed for the interaction with an IC50 of ∼700 nM (n = 3, error bars, s.d.).
Extended Data Figure 2 Purification and crystallization of T4L–rhodopsin–arrestin.
a, Purification of the T4L–rhodopsin–arrestin (T4L–Rho–Arr) complex. His8–MBP–MBP–T4L–Rho–Arr complex was first purified by amylose column chromatography (lane 1). The His8–MBP–MBP tandem tag was then released by cleavage with 3C protease (lane 2) and removed by binding to Ni-NTA beads to recover pure T4L–rhodopsin–arrestin (T4L–Rho–Arr) protein (lane 3). b, Analytical gel filtration profile of the T4L–rhodopsin–arrestin complex. T4L–rhodopsin–arrestin eluted mostly as monomers with a small proportion of oligomers. The molecular weights of protein standards are indicated at the top. c, Thermal stability shift analysis of T4L–rhodopsin–arrestin. T4L–rhodopsin–arrestin is relatively stable with a Tm of 59 °C. d, e, Crystals of T4L–rhodopsin–arrestin in lipid cubic phase under bright-field illumination (d) and polarized light (e). f, X-ray diffraction pattern of a T4L–rhodopsin–arrestin crystal recorded at LS-CAT of APS. The green ring indicates the position of reflections at 8.0 Å resolution.
Extended Data Figure 3 Electron density map for the overall complex and the key interfaces based on the XFEL data.
a, A 2Fo − Fc electron density map contoured at 1σ of the arrestin finger loop, which forms the key interface with TM7 and helix 8. b, A 2Fo − Fc electron density map contoured at 1σ of the loop between TM5 and TM6, which forms the key interface with the β-strand following the finger loop. c, A 3,000 K simulated annealing omit map (2Fo − Fc electron density map contoured at 1σ) calculated from the 3.8 Å/3.8 Å/3.3 Å XFEL data supports the overall arrangement of the rhodopsin–arrestin complex. In all panels, the complex structure is shown with rhodopsin coloured in green and arrestin in brown. d, The C-loop with a 2Fo − Fc composite omit map at 1σ calculated from the 3.8 Å/3.8 Å/3.3 Å truncated XFEL data. Key residues are labelled.
Extended Data Figure 4 Structure similarity of the four rhodopsin–arrestin complexes in the asymmetric units and the interface between rhodopsin and arrestin.
a, Two 90° views of the superposition of the four rhodopsin–arrestin complexes are shown as cartoon representation. The four complexes have an r.m.s.d. of less than 0.5 Å in the Cα atoms of rhodopsin and arrestin. b, Close-up view of arrestin-binding sites in rhodopsin. The four arrestin-binding sites (P1–P4) are highlighted in brown on the rhodopsin surface. The rhodopsin C-terminal tail/arrestin interface (P4) is based on computational modelling and disulfide cross-linking data. c, Rhodopsin-binding sites in arrestin. The four rhodopsin-binding sites (P1–P4) are highlighted in green on the arrestin surface.
Extended Data Figure 5 Conformational modelling of the rhodopsin–arrestin full length complex.
a, An overview of the computational model. b, Predicted interactions of the rhodopsin C terminus with arrestin, showing strong to medium pairwise restraints between Cβ atoms of rhodopsin and arrestin residues identified by disulfide crosslinking. c, Same as in b, but showing predicted hydrogen bonding and ionic interactions for the C-terminal residues of rhodopsin.
Extended Data Figure 6 Dynamics of free 3A arrestin and rhodopsin-bound arrestin determined by HDX.
a, HDX perturbation map between rhodopsin-bound arrestin and free arrestin, which is derived from the difference in the HDX rate between rhodopsin-bound arrestin and free arrestin. The bars below the arrestin sequence represent the peptide fragments resolved by mass spectrometry and the colours of the bars indicate the relative decrease in deuterium exchange (colour code at bottom). b, The thermal stability of free 3A arrestin and the rhodopsin–arrestin complex shows that the rhodopsin–arrestin complex is more stable than free 3A arrestin.
Extended Data Figure 7 Cell-based Tango assays to validate the rhodopsin–arrestin interface.
a, Cartoon illustration of the Tango assay for rhodopsin–arrestin interactions in cells. b, c, Mutations of key arrestin (b) and rhodopsin (c) residues that mediate the rhodopsin–arrestin interactions. Tango assay were performed in the absence or presence of 10 µM all-trans-retinal (ATR). (n = 3, error bars, s.d.).
Extended Data Figure 8 Control experiments for disulfide bond cross-linking specificity.
a, The product of the cross-linking reaction of finger loop residue G77C with N3107.57C of TM7 was confirmed by western blots using anti-Flag antibody (which detects arrestin–Flag fusion) and anti-HA antibody (which detects rhodopsin–HA fusion). The cross-linked products are marked with arrow heads, and free-arrestin and free-rhodopsin are indicated by asterisks. Arrestin (3A) and rhodopsin (4M) without cysteine mutations do not form cross-linked products. b, The cross-linked product of finger loop residue G77C with N3107.57C of TM7 was sensitive to treatment with reducing agents, indicating the cross-linking is mediated through disulfide bond formation. c, A close-up view of arrestin finger loop residues M76C and G77C and their cross-linking with rhodopsin, which shows that G77C was specifically cross-linked to N3107.57C of TM7 and Q3128.49 of helix 8, and M76C was cross-linked to N3107.57C of TM7 and Q3128.49C of helix 8, but not to other residues. d, Structure and cross-linking of finger loop N-terminal residues Q70C, E71C, and D72C of arrestin to T70C and K67C from ICL1 of rhodopsin. e, Structure and cross-linking of arrestin back loop residues R319C and T320C to Q237ICL3C from TM5 of rhodopsin.
Extended Data Figure 9 Structure comparison of the arrestin-bound rhodopsin with the β2-adrenergic receptor in complex with Gs protein (PDB code 3SN6) and the inactive rhodopsin (PDB code 1F88).
a, Superposition of arrestin-bound rhodopsin (green) with Gs protein-bound β2 adrenergic receptor (light yellow). The major conformational changes are indicated by arrows. b, An intracellular view of a superposition of arrestin-bound rhodopsin (green) and Gs protein-bound β2-adrenergic receptor (light yellow). c, Overlays of arrestin-bound rhodopsin (green) with inactive rhodopsin (pink) reveals specific conformational changes in each TM helix. The arrows indicate outward movements of TM helices. d, r.m.s.d. of Cα atom differences between arrestin-bound rhodopsin and inactive rhodopsin shows the large conformational changes in TM5 and TM6.
Extended Data Figure 10 Structure of rhodopsin-bound arrestin and its comparison with inactive and ‘pre-activated’ arrestin.
a, b, The charge potential surface map of rhodopsin from the rhodopsin–arrestin bound complex shows that the cytoplasmic rhodopsin TM bundle surface is positively charged (blue) whereas its C-terminal tail is negatively charged (red). c, d, Charged surface of arrestin from the rhodopsin–arrestin bound complex shows that the arrestin finger loop is negatively charged (red) and its N-terminal β-strand interface is positively charged (blue). The charge distribution in rhodopsin and arrestin is complementary to each other for their interactions. e, Comparison of rhodopsin-bound arrestin (light blue) with inactive arrestin (brown, PDB code: 1CF1), showing an ∼20° rotation between the N- and C- domains of arrestin. f, Comparison of rhodopsin-bound arrestin (dark brown) with pre-activated arrestin (light brown, PDB code 4J2Q), showing conformational changes in the finger loop, which adopts an α-helical conformation (cyan) in the complex. The extended finger loop conformation would protrude into the rhodopsin TM bundle and is not compatible with receptor binding. Computational model for the full rhodopsin–arrestin complex is shown in panels b and d.
Extended Data Figure 11 A computational model of phosphorylated rhodopsin in complex with arrestin and salt sensitivity of the rhodopsin–arrestin interaction.
a–d, An overall view (a) and close-up views (b–d) of the computational model of the rhodopsin C-tail with phospho-serine at positions 334, 338 and 343 in complex with arrestin. e, The AlphaScreen control (biotin–His6) shows much less salt sensitivity than the interaction between His-tag–rhodopsin and biotin arrestin, which is very sensitive to salt, with an IC50 of around 200 mM NaCl (100 mM NaCl added to 100 mM salt of the original assay buffer) (n = 3, error bars, s.d.).
Extended Data Figure 12 A positive charge property is commonly found at the cytoplasmic side of GPCRs.
a–e, Surface charge potential of the cytoplasmic side of selected agonist bound GPCR structures: β1AR, PDB code 2Y02 (a); β2AR, PDB code 3PDS (b); A2A adenosine receptor, PDB code 3QAK (c); serotonin receptor 5HT1B, PDB code 4IAR (d); serotonin receptor 5HT2B, PDB code 4IB4 (e). Positive and negative charge potentials are shown in blue and red, respectively. f, Sequence alignment of the finger loop region highlighting negatively charged residues (shown in red), which are conserved in all subtypes of arrestins.
Extended Data Figure 13 A possible role of the arrestin C-edge in lipid binding.
a, b, The asymmetric assembly of the rhodopsin–arrestin complex in the presence of a lipid membrane bilayer, showing the C-edge of arrestin dipping into the lipid layer. c, d, A close-up view of the C-edge of arrestin in the membrane layer, where the conserved hydrophobic side chains are shown. The figure was made using the computational model for the full rhodopsin–arrestin complex.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-5. (PDF 306 kb)
Rights and permissions
About this article
Cite this article
Kang, Y., Zhou, X., Gao, X. et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523, 561–567 (2015). https://doi.org/10.1038/nature14656
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14656
This article is cited by
-
Dynamic Nature of Proteins is Critically Important for Their Function: GPCRs and Signal Transducers
Applied Magnetic Resonance (2024)
-
Structure, function and drug discovery of GPCR signaling
Molecular Biomedicine (2023)
-
Sequestration of Gβγ by deubiquitinated arrestins into the nucleus as a novel desensitization mechanism of G protein–coupled receptors
Cell Communication and Signaling (2023)
-
Structural details of a Class B GPCR-arrestin complex revealed by genetically encoded crosslinkers in living cells
Nature Communications (2023)
-
Computational investigation of functional water molecules in GPCRs bound to G protein or arrestin
Journal of Computer-Aided Molecular Design (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.