Abstract
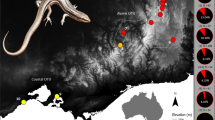
Sex determination in animals is amazingly plastic. Vertebrates display contrasting strategies ranging from complete genetic control of sex (genotypic sex determination) to environmentally determined sex (for example, temperature-dependent sex determination)1. Phylogenetic analyses suggest frequent evolutionary transitions between genotypic and temperature-dependent sex determination in environmentally sensitive lineages, including reptiles2. These transitions are thought to involve a genotypic system becoming sensitive to temperature, with sex determined by gene–environment interactions3. Most mechanistic models of transitions invoke a role for sex reversal3,4,5. Sex reversal has not yet been demonstrated in nature for any amniote, although it occurs in fish6 and rarely in amphibians7,8. Here we make the first report of reptile sex reversal in the wild, in the Australian bearded dragon (Pogona vitticeps), and use sex-reversed animals to experimentally induce a rapid transition from genotypic to temperature-dependent sex determination. Controlled mating of normal males to sex-reversed females produces viable and fertile offspring whose phenotypic sex is determined solely by temperature (temperature-dependent sex determination). The W sex chromosome is eliminated from this lineage in the first generation. The instantaneous creation of a lineage of ZZ temperature-sensitive animals reveals a novel, climate-induced pathway for the rapid transition between genetic and temperature-dependent sex determination, and adds to concern about adaptation to rapid global climate change.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Sarre, S. D., Georges, A. & Quinn, A. The ends of a continuum: genetic and temperature-dependent sex determination in reptiles. Bioessays 26, 639–645 (2004)
Sarre, S. D., Ezaz, T. & Georges, A. Transitions between sex-determining systems in reptiles and amphibians. Annu. Rev. Genom. Hum. Genet. 12, 391–406 (2011)
Bull, J. J. Evolution of environmental sex determination from genotypic sex determination. Heredity 47, 173–184 (1981)
Quinn, A. E., Sarre, S. D., Ezaz, T., Graves, J. A. M. & Georges, A. Evolutionary transitions between mechanisms of sex determination in vertebrates. Biol. Lett. 7, 443–448 (2011)
Schwanz, L. E., Ezaz, T., Gruber, B. & Georges, A. Novel evolutionary pathways of sex-determining mechanisms. J. Evol. Biol. 26, 2544–2557 (2013)
Chan, S. T. H. & Yeung, W. S. B. Sex control and sex reversal in fish under natural conditions. Fish Physiol. 9, 171–222 (1983)
Alho, J. S., Matsuba, C. & Merila, J. Sex reversal and primary sex ratios in the common frog (Rana temporaria). Mol. Ecol. 19, 1763–1773 (2010)
Miura, I. Sex chromosome differentiation in the Japanese brown frog, Rana japonica. I. Sex-related heteromorphism of the distribution pattern of constitutive heterochromatin in chromosome no. 4 of the Wakuya population. Zool. Sci. 11, 797–806 (1994)
Bachtrog, D. et al. Sex determination: why so many ways of doing it? PLoS Biol. 12, e1001899 (2014)
Shine, R., Elphick, M. J. & Donnellan, S. Co-occurrence of multiple, supposedly incompatible modes of sex determination in a lizard population. Ecol. Lett. 5, 486–489 (2002)
Radder, R. S., Quinn, A. E., Georges, A., Sarre, S. D. & Shine, R. Genetic evidence for co-occurrence of chromosomal and thermal sex-determining systems in a lizard. Biol. Lett. 4, 176–178 (2008)
Quinn, A. E. et al. Temperature sex reversal implies sex gene dosage in a reptile. Science 316, 411 (2007)
Mork, L., Czerwinski, M. & Capel, B. Predetermination of sexual fate in a turtle with temperature-dependent sex determination. Dev. Biol. 386, 264–271 (2014)
Wang, C. et al. Identification of sex chromosomes by means of comparative genomic hybridization in a lizard, Eremias multiocellata. Zool. Sci. 32, 151–156 (2015)
Ezaz, T. et al. The dragon lizard Pogona vitticeps has ZZ/ZW micro-sex chromosomes. Chromosome Res. 13, 763–776 (2005)
Quinn, A. E., Ezaz, T., Sarre, S. D., Graves, J. A. M. & Georges, A. Extension, single-locus conversion and physical mapping of sex chromosome sequences identify the Z microchromosome and pseudo-autosomal region in a dragon lizard, Pogona vitticeps. Heredity 104, 410–417 (2010)
Ezaz, T. et al. Sequence and gene content of a large fragment of a lizard sex chromosome and evaluation of candidate sex differentiating gene R-spondin 1. BMC Genom. 14, 899 (2013)
Booth, W. et al. Facultative parthenogenesis discovered in wild vertebrates. Biol. Lett. 8, 983–985 (2012)
Booth, W., Johnson, D. H., Moore, S., Schal, C. & Vargo, E. L. Evidence for viable, non-clonal but fatherless boa constrictors. Biol. Lett. 7, 253–256 (2011)
Watts, P. C. et al. Parthenogenesis in Komodo dragons. Nature 444, 1021–1022 (2006)
Charnov, E. L. & Bull, J. When is sex environmentally determined? Nature 266, 828–830 (1977)
Warner, D. A. & Shine, R. The adaptive significance of temperature-dependent sex determination in a reptile. Nature 451, 566–568 (2008)
Warner, D. A. & Shine, R. The adaptive significance of temperature-dependent sex determination: experimental tests with a short lived lizard. Evolution 59, 2209–2221 (2005)
McGaugh, S. E. & Janzen, F. J. Effective heritability of targets of sex-ratio selection under environmental sex determination. J. Evol. Biol. 24, 784–794 (2011)
Saunders, P. A. et al. XY females do better than the XX in the african pygmy mouse, Mus minutoides. Evolution 68, 2119–2127 (2014)
Bull, J. J. Evolution of Sex Determining Mechanisms (Benjamin-Cummings, 1983)
Sinervo, B. et al. Erosion of lizard diversity by climate change and altered thermal niches. Science 328, 894–899 (2010)
Hoffmann, A. A. & Sgro, C. M. Climate change and evolutionary adaptation. Nature 470, 479–485 (2011)
Boyle, M., Hone, J., Schwanz, L. E. & Georges, A. Under what conditions do climate-driven sex ratios enhance versus diminish population persistence? Ecol. Evol. 4, 4522–4533 (2014)
Mrosovsky, N. & Pieau, C. Transitional range of temperature, pivotal temperatures and thermosensitive stages for sex determination in reptiles. Amphib.-Reptil. 12, 169–179 (1991)
Harlow, P. S. A harmless technique for sexing hatchling lizards. Herpetol. Rev. 27, 71–72 (1996)
Brown, D. Hemipenal transillumination as a sexing technique in varanids. Biawak 3, 26–29 (2009)
Ezaz, T. et al. A simple non-invasive protocol to establish primary cell lines from tail and toe explants for cytogenetic studies in Australian dragon lizards (Squamata: Agamidae). Cytotechnology 58, 135–139 (2008)
Young, M. J., O’Meally, D., Sarre, S. D., Georges, A. & Ezaz, T. Molecular cytogenetic map of the central bearded dragon, Pogona vitticeps (Squamata: Agamidae). Chromosome Res. 21, 361–374 (2013)
Sumner, A. T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 75, 304–306 (1972)
Matsubara, K. et al. Highly differentiated ZW sex microchromosomes in the Australian Varanus species evolved through rapid amplification of repetitive sequences. PLoS ONE 9, e95226 (2014)
Cioffi, M. B., Martins, C., Vicari, M. R., Rebordinos, L. & Bertollo, L. A. Differentiation of the XY sex chromosomes in the fish Hoplias malabaricus (Characiformes, Erythrinidae): unusual accumulation of repetitive sequences on the X chromosome. Sex Dev. 4, 176–185 (2010)
Courtois, B. et al. Genome-wide association mapping of root traits in a japonica rice panel. PLoS ONE 8, e78037 (2013)
Cruz, V. M., Kilian, A. & Dierig, D. A. Development of DArT marker platforms and genetic diversity assessment of the U.S. collection of the new oilseed crop lesquerella and related species. PLoS ONE 8, e64062 (2013)
Kilian, A. et al. Diversity arrays technology: a generic genome profiling technology on open platforms. Methods Mol. Biol. 888, 67–89 (2012)
Raman, H. et al. Genome-wide delineation of natural variation for pod shatter resistance in Brassica napus. PLoS ONE 9, e101673 (2014)
Acknowledgements
We thank the following persons and institutions: W. Ruscoe and J. Richardson (animal husbandry); A. Quinn, M. Young and J. Richardson (field collections); A. Kilian and Diversity Arrays Technology (SNP genotyping); A. Livernois (sequencing assistance); B. Gruber and N. Garlapati (geographic information system); A. T. Adamack (modelling advice); A. Dobos and P. E. Geertz (graphic design); J. Deakin, R. Thompson and the Kioloa Science Writers Workshop (revisions of the manuscript). We particularly thank reviewer J. J. Bull for suggesting the modelling approach in Extended Data Fig. 4. Funding was from Australian Research Council Discovery Grant DP110104377 to A.G. and T.E. This research was conducted under appropriate approvals from the Victorian, New South Wales and Queensland authorities, and with approvals from the Animal Ethics Committee of the University of Canberra.
Author information
Authors and Affiliations
Contributions
C.E.H. and A.G. designed the study. C.E.H. conducted breeding experiments, egg incubations, parentage SNP analysis and prepared figures. D.O'M. collected the animals from the field. C.E.H. and X.Z. conducted the molecular sex testing. B.A. and K.M. undertook the cytogenetic analysis and prepared extended data figures, under the supervision of T.E. A.G. and C.E.H. undertook the statistical analyses and A.G. conducted the modelling of ZW genotype frequency with temperature. All authors contributed to writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 C-banded P. vitticeps chromosomes.
a, Mitotic metaphase chromosomes of a ZW control female individual. Arrowhead indicates the presence of a W chromosome identified by dense black staining of a single microchromosome. b, Mitotic metaphase chromosomes of a female putative ZZ sex-reversed individual. No evidence of a W chromosome was detected. c, Mitotic metaphase chromosomes of a control ZZ male individual. No evidence of a W chromosome was detected. Scale bar, 10 μm.
Extended Data Figure 2 Comparative genomic hybridization in P. vitticeps.
Genomic DNA was labelled by nick translation incorporating SpectrumGreen-dUTP for males and SpectrumOrange-dUTP for females. a, Mitotic metaphase chromosomes of a ZW control female individual. Arrowhead indicates the presence of a single W microchromosome identified by the enriched orange fluorescence of female specific genomic DNA labelled with SpectrumOrange-dUTP. b, Mitotic metaphase chromosomes of a female putative ZZ sex-reversed individual. No evidence of a W chromosome was detected. c, Mitotic metaphase chromosomes of a control ZZ male individual. No evidence of a W chromosome was detected. d–f, DAPI staining of the same metaphases, control ZW female, sex-reversed ZZ female and control ZZ male, respectively. Scale bar, 10 μm.
Extended Data Figure 3 Physical mapping of a W-chromosome-linked microsatellite motif in P. vitticeps.
a, Mitotic metaphase chromosomes of a ZW control female individual. Arrowhead indicates the presence of a W chromosome identified by a strong hybridization of (AAGG)8-Cy3 florescence (orange) on a single microchromosome. b, Mitotic metaphase chromosomes of a female putative ZZ sex-reversed individual. No evidence of a W chromosome was detected. c, Mitotic metaphase chromosomes of a control ZZ male individual. No evidence of a W chromosome was detected. d–f, DAPI staining of the same metaphases, control ZW female, sex-reversed ZZ female and control ZZ male, respectively. Scale bar, 10 μm.
Extended Data Figure 4 Modelling the decline of the ZW genotype resulting from frequency-dependent selection.
Frequency of the ZW genotype declines precipitously with increasing incubation temperature. Our wild population (shown in red, 14.3% sex reversal) resides on the precipice between GSD and TSD and requires only a small change in environmental temperature to precipitate loss of the W chromosome.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-4. (PDF 169 kb)
Supplementary data
This file contains the source data used to make Supplementary Table 1. (XLSX 1099 kb)
Rights and permissions
About this article
Cite this article
Holleley, C., O'Meally, D., Sarre, S. et al. Sex reversal triggers the rapid transition from genetic to temperature-dependent sex. Nature 523, 79–82 (2015). https://doi.org/10.1038/nature14574
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14574
This article is cited by
-
Gene expression of male pathway genes sox9 and amh during early sex differentiation in a reptile departs from the classical amniote model
BMC Genomics (2023)
-
Developmental dynamics of sex reprogramming by high incubation temperatures in a dragon lizard
BMC Genomics (2022)
-
Effects of incubation temperature on development, morphology, and thermal physiology of the emerging Neotropical lizard model organism Tropidurus torquatus
Scientific Reports (2022)
-
Sex determination mechanisms and sex control approaches in aquaculture animals
Science China Life Sciences (2022)
-
Dispersal and mating patterns determine the fate of naturally dispersed populations: evidence from Bombina orientalis
BMC Ecology and Evolution (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.