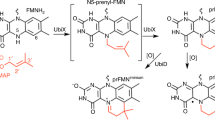
Abstract
Ubiquinone (also known as coenzyme Q) is a ubiquitous lipid-soluble redox cofactor that is an essential component of electron transfer chains1. Eleven genes have been implicated in bacterial ubiquinone biosynthesis, including ubiX and ubiD, which are responsible for decarboxylation of the 3-octaprenyl-4-hydroxybenzoate precursor2. Despite structural and biochemical characterization of UbiX as a flavin mononucleotide (FMN)-binding protein, no decarboxylase activity has been detected3,4. Here we report that UbiX produces a novel flavin-derived cofactor required for the decarboxylase activity of UbiD5. UbiX acts as a flavin prenyltransferase, linking a dimethylallyl moiety to the flavin N5 and C6 atoms. This adds a fourth non-aromatic ring to the flavin isoalloxazine group. In contrast to other prenyltransferases6,7, UbiX is metal-independent and requires dimethylallyl-monophosphate as substrate. Kinetic crystallography reveals that the prenyltransferase mechanism of UbiX resembles that of the terpene synthases8. The active site environment is dominated by π systems, which assist phosphate-C1′ bond breakage following FMN reduction, leading to formation of the N5–C1′ bond. UbiX then acts as a chaperone for adduct reorientation, via transient carbocation species, leading ultimately to formation of the dimethylallyl C3′–C6 bond. Our findings establish the mechanism for formation of a new flavin-derived cofactor, extending both flavin and terpenoid biochemical repertoires.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lenaz, G. & Genova, M. L. Mobility and function of coenzyme Q (ubiquinone) in the mitochondiral respiratory chain. Biochim. Biophys. Acta 1787, 563–573 (2009)
Aussel, L. et al. Biosynthesis and physiology of coenzyme Q in bacteria. Biochim. Biophys. Acta 1837, 1004–1011 (2014)
Gulmezian, M., Hyman, K. R., Marbois, B. N., Clarke, C. F. & Javor, G. T. The role of UbiX in Escherichia coli coenzyme Q biosynthesis. Arch. Biochem. Biophys. 467, 144–153 (2007)
Rangarajan, E. S. et al. Crystal structure of a dodecameric FMN-dependent UbiX-like decarboxylate (Pad1) from Eschericia coli O157:H7. Protein Sci. 13, 3006–3016 (2004)
Payne, K. A. P. et al. New cofactor supports α,β-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition. Nature http://www.dx.doi.org/10.1038/nature14560 (2015)
Liang, P. H. Reaction kinetics, catalytic mechanisms, conformational changes, and inhibitor design for prenyltransferases. Biochemistry 48, 6562–6570 (2009)
Doud, E. H., Perlstein, D. L., Wolpert, M., Cane, D. E. & Walker, S. Two distinct mechanisms for TIM barrel prenyltransferases in bacteria. J. Am. Chem. Soc. 133, 1270–1273 (2011)
Gao, Y. & Honzatko, R. B. Peters, terpenoid synthase structures: a so far complete view of complex catalysis. Nat. Prod. Rep. 29, 1153–1175 (2012)
Walsh, C. T. & Wencewicz, T. A. Flavoenzymes: versatile catalysts in biosynthetic pathways. Nat. Prod. Rep. 30, 175–200 (2013)
Heuts, D. P., Scrutton, N. S., McIntire, W. S. & Fraaije, M. W. What’s in a covalent bond? On the role and formation of covalently bound flavin cofactors. FEBS J. 276, 3405–3427 (2009)
Mukai, N., Masaki, K., Fujii, T., Kawamukai, M. & Iefuji, H. PAD1 and FDC1 are essential for the decarboxylation of phenylacrylic acids in Saccharomyces cerevisiae . J. Biosci. Bioeng. 109, 564–569 (2010)
Lin, F., Ferguson, K. L., Boyer, D. R., Lin, X. N. & Marsh, E. N. Isofunctional enzymes Pad1 and UbiX catalyse formation of a novel cofactor required by ferulic acid decarboxylase and 4-hydroxy-3-polyprenylbenzoic acid decarboxylase. ACS Chem. Biol. 10, 1137–1144 (2015)
Kopec, J., Schnell, R. & Schneider, G. Structure of PA4019, a putative aromatic acid decarboxylase from Pseudomonas aeruginosa . Acta Crystallogr. F 67, 1184–1188 (2011)
Fraaije, M. W. & Mattevi, A. Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem. Sci. 25, 126–132 (2000)
Berthelot, K., Estevez, Y., Deffieux, A. & Peruch, F. Isopentenyl diphosphate isomerase: a checkpoint to isoprenoid biosynthesis. Biochimie 94, 1621–1634 (2012)
Nagai, T. et al. Covalent modification of reduced flavin mononucleotide in type-2 isopentenyl diphosphate isomerase by active-site-directed inhibitors. Proc. Natl Acad. Sci. USA 108, 20461–20466 (2011)
Christianson, D. W. Unearthing the roots of the terpenome. Curr. Opin. Chem. Biol. 12, 141–150 (2008)
Smanski, M. J., Peterson, R. M., Huang, S. X. & Shen, B. Bacterial diterpene synthases: new opportunities for mechanistic enzymology and engineered biosynthesis. Curr. Opin. Chem. Biol. 16, 132–141 (2012)
Lupa, B., Lyon, D., Gibbs, M. D., Reeves, R. A. & Wiegel, J. Distribution of genes encoding the microbial non-oxidative reversible hydroxyarylic acid decarboxylates/phenol carboxylases. Genomics 86, 342–351 (2005)
Abu Laban, N., Selesi, D., Rattie, T., Tischler, P. & Meckenstock, R. U. Identification of enzymes involved in anaerobic benzene degradation by a strictly anaerobic iron-reducing enrichment culture. Environ. Microbiol. 12, 2783–2796 (2010)
Erb, T. J. Carboxylases in natural and synthetic microbial pathways. Appl. Environ. Microbiol. 77, 8466–8477 (2011)
Chang, W. C., Song, H., Liu, H. W. & Liu, P. Current development in isoprenoid precursor biosynthesis and regulation. Curr. Opin. Chem. Biol. 17, 571–579 (2013)
VanNice, J. C. et al. Identification in Haloferax volcanii of phosphomevalonate decarboxylase and isopentenyl phosphate kinase as catalysts of the terminal enzyme reactions in an archaeal alternate mevalonate pathway. J. Bacteriol. 196, 1055–1063 (2014)
Sobrado, P. Noncanonical reactions of flavoenzymes. Int. J. Mol. Sci. 13, 14219–14242 (2012)
Sun, H. G., Ruszczycky, M. W., Chang, W. C., Thibodeaux, C. J. & Liu, H. W. Nucleophilic participation of reduced flavin coenzyme in mechanism of UDP-galactopyranose mutase. J. Biol. Chem. 287, 4602–4608 (2012)
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Fu, G. et al. Atomic-resolution structure of an N5 flavin adduct in d-arginine dehydrogenase. Biochemistry 50, 6292–6294 (2011)
Frisch, X. M. J. et al. Gaussian 09 (Gaussian, Wallingford, Connecticut, revision B.01, 2010)
Guex, Y. N. & Peitsch, M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 (1997)
Acknowledgements
This work was supported by BBSRC grants (BB/K017802/1 with Shell and BB/M017702/1). We thank Diamond Light Source for access to beamlines (proposal number MX8997) that contributed to the results presented here. S.H. is a BBSRC David Phillips research fellow. N.S.S. is an EPSRC Established Career Fellow and Royal Society Wolfson Award holder. The authors acknowledge the assistance given by IT Services and the use of the Computational Shared Facility and the Protein Structure Facility at The University of Manchester.
Author information
Authors and Affiliations
Contributions
M.D.W. carried out molecular biology, biophysical and structural biology studies together with K.A.P.P. and D.L. M.D.W. and S.A.M. performed in vitro reconstitution experiments. K.F. and S.E.J.R. performed and analysed EPR experiments. S.H. performed DFT calculations. N.J.W.R. and D.K.T. undertook liquid chromatography–mass spectrometry of extracts and with R.G. interpreted the data on substrate–product species. All authors discussed the results with D.P and N.S.S. and participated in writing the manuscript. D.L. initiated and directed this research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Mass spectrometric analysis of the UbiX product.
a Structural elucidation of the reduced UbiX product. From an initial full scan TIC on UbiX extract, a 525 m/z ion extracted chromatogram was created under a gradient elution using H2O/acetonitrile both containing 0.1% formic acid indicating a major peak apex at 9.57 min with a 54/46 solvent elution composition (not shown). Subsequent data dependant TIC and 525 m/z scan extracted chromatograms were created under 55% A/45% B isocratic solvent elution and ion extraction between 524.5–525.5 m/z produced a singular peak at 2.28 min displaying an associated full-scan molecular ion peak with m/z = 525.1726 (M+ = C22H30N4O9P) at a resolution of 58,500 with a mass accuracy of 3.59 p.p.m. Fragmentation of the 525.1726 m/z molecular ion peak in an automated data dependent manner using helium-based chemical-induced dissociation (CID level 35) generated a spectral tree that indicates the removal of the newly formed, more labile, tertiary ring at the MS2 level. Subsequent removal of the phosphate head group at the MS3 level was achieved using CID 35 on the 456.23 m/z molecular species with a final MS4 step using CID 35 on 358.18 m/z completely removing the tail group from the central three-ring isoalloxazine system. b, Structural elucidation of the re-oxidized UbiX/Fdc1 cofactor. From an initial full scan TIC on UbiX extract (i), a 524 m/z ion extracted chromatogram was created under a gradient elution using H2O/acetonitrile both containing 0.1% formic acid indicating a major peak apex at 9.24 min with a 48/52 solvent composition (not shown). Subsequent data dependant TIC and 524 m/z scan extracted chromatograms (ii) were created under 50% A/50% B isocratic solvent elution and ion extraction between 523.5–524.5 m/z produced a singular peak at 2.08 min displaying an associated full-scan molecular ion peak with m/z = 524.1656 (M+ = C22H29N4O9P) at a resolution of 58,500 with a mass accuracy of 2.78 p.p.m. Fragmentation of the 524.1656 m/z molecular ion peak in an automated data dependent manner using helium-based chemical-induced dissociation (CID level 35) generated a spectral tree (iii) that indicates the removal of the newly formed, more labile, tertiary ring at the MS2 level. Subsequent removal of the phosphate head group at the MS3 level was achieved using CID 35 on the 498.31 m/z molecular species to create 327.18 (A) alongside a sister fragment 455.31 (B) that represents the full removal of the tertiary ring but retaining the phosphate head group.
Extended Data Figure 2 EPR spectroscopic analysis of the UbiX radical product.
a, X-band continuous wave EPR spectra of UbiX in frozen solution: (i) wild type (WT) as isolated; (ii) WT plus DMAP; (iii) WT reduced with dithionite; (iv) WT + DMAP reduced with dithionite; (v) WT + DMAP reduced with dithionite and reoxidized with oxygen; (vi) Y169F mutant + DMAP reduced with dithionite and reoxidized with oxygen; (vii) W200F mutant + DMAP reduced with dithionite and reoxidized with oxygen. The FMN–DMAP adduct radical is only formed when UbiX is reoxidized in the presence of DMAP and this formation is not affected by mutation of those aromatic residues forming the π-cage that could give rise to Y or W radical species. b, X-band continuous wave EPR spectra of frozen solutions of WT UbiX + DMAP and reduced with dithionite with the addition of potassium ferricyanide to the following concentrations: (i) 260 μM; (ii) 160 μM; (iii) 50 μM; (iv) 40 μM; (v) 30 μM; (vi) 20 μM; (vii) 0 μM. Experimental conditions: microwave power 10 μW, modulation amplitude 1.5 G, temperature 20 K. Showing the radical can also be formed using chemical oxidation in the absence of oxygen and thus does not arise from a peroxide species generated by the reaction of reduced oxygen species formed when the dithionite sample is exposed to oxygen. An initial radical is formed under these conditions that exhibits a considerably broader EPR signal than the prFMNradical and is as yet unidentified.
Extended Data Figure 3 Pulsed Davies ENDOR spectra of the prFMNradical–UbiX complex.
The spectrum was measured at a field equivalent to gav = 2.0033. Although a complete assignment of the spectrum requires specific deuteration of FMN and DMAP, the ENDOR spectrum is dominated by two large hyperfine couplings to β-protons indicated as HA and HB. Using the Heller-McConnell equation the values of the dihedral angles, θ, can be determined as shown and are consistent with the orientation of the C1′-protons of the DMAP-derived fragment of the radical observed crystallographically, as shown in the figure above. The unpaired electron spin density, ρ, at N5 of the FMN-derived fragment of the radical can also be estimated from the Heller-McConnell equation. B′ is negligible, whereas B′′ is thought to have a value of ∼160, although studies of β-protons coupled to unpaired electron spin density at a nitrogen atom are rare, giving an unpaired spin density at N5 of ∼0.3, consistent with calculations and considerably smaller than the unpaired electron spin density of 0.4 or greater expected for C1′ of an aromatic amino acid radical.
Extended Data Figure 4 DFT modelling of the UbiX product.
Top, DFT model of the purple radical species showing the location of significant atomic spin densities (>|0.02|) to the right. The optimized structure (blue carbons) overlaid with the crystal coordinates (green carbons) is shown below. The model was geometry optimized in the gas phase using the UB3LYP/6-311++G(d,p) level of theory. Cartesian coordinates of the optimized structure are given in the Supplementary Information.
Extended Data Figure 5 Additional UbiX solution studies.
a, Reconstitution of A. niger Fdc1 activity with UbiX–prFMNreduced and prFMNreduced obtained through filtration of a UbiX–prFMNreduced reaction. Control reactions are devoid of any DMAP substrate. b, Rate of formation of spectral species 2 (see Fig. 1f) in function of DMAP concentration. c, Rate of decay of spectral species 2 (see Fig. 1f) in function of DMAP concentration. d, Spectral species obtained from singular value decomposition of rapid-scan stopped-flow spectrophotometric data following mixing of UbiX–prFMNreduced with oxygenated buffer. e, The rate of purple radical (species B in panel d of this figure) formation as obtained from singular value decomposition of rapid-scan stopped-flow spectrophotometric data following mixing of UbiX–prFMNreduced with oxygenated buffer has a linear dependence on oxygen concentration. Error bars are s.e.m., n = 3.
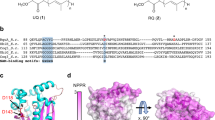
Extended Data Figure 6 Multiple sequence alignment of UbiX/Pad1 enzymes from selected bacterial or fungal species.
Pseudomonas aeruginosa UbiX (NP_252708), Escherichia coli O157:H7 EcdB (NP_311620), Escherichia coli UbiX (YP_490553), Bacillus subtilis BsdB (WP_009966530), Saccharomyces cerevisiae Pad1 (AAB64980), Aspergillus niger PadA1 (ABN13117), and orf8 from the Thauera aromatica phenylphosphate carboxylase gene cluster (PAAD_THAAR). Conserved residues involved in phosphate binding, N5 polar network or formation of the substrate binding π-cage are indicated by labelled arrows. Secondary structure elements of P. aeruginosa UbiX crystal structure are shown. Alpha helices and 310 helices (denoted as η) are shown as squiggles, β-strands as arrows and β-turns as TT.
Extended Data Figure 7 Crystal structure of P. aeruginosa UbiX–FMN–DMAP flash cooled to 100 K at 30 s following complete reduction by sodium dithionite.
Two orientations are displayed as in Fig. 2. The omit map for the prFMNreduced product is shown as a green mesh, contoured at 4σ.
Extended Data Figure 8 Crystal structures of P. aeruginosa UbiXY169F.
a, Detailed view of the UbiXY169–FMN–DMAP complex with individual amino acids contributing to active site structure shown in atom-coloured sticks (carbons colour coded as in Fig. 2a). Two orientations are displayed as in Fig. 2. The omit map for the DMAP substrate is shown as green mesh, contoured at 4σ. b, Detailed view of the UbiXY169F N5–C1′ adduct species obtained through flash-cooling following reduction. The omit map for the N5–C1′ adduct is shown as a green mesh, contoured at 4σ.
Extended Data Figure 9 DFT models of proposed intermediate species in the UbiX reaction.
a, DFT models of species II and IVa (as defined in Fig. 4). Conversion from II to IVa is achieved by ∼180° rotation about C1′–C2′ (blue arrow) and the N5–H and methanol species (red) are only found in species IVa models. b, Overlay of the species II DFT model (green carbons) with the crystal coordinates of species II and S15 (teal carbons). c, Three DFT models of IVa were examined and two orthogonal projections are shown overlaid with the crystal coordinates (teal carbons): (Vi, yellow carbons) with a methanol analogue of S15 (a, in red) with the C–N5 distance fixed to the crystallographic distance of 4.0 Å; (Vii, magenta carbons) with N5 protonated (no methanol), and (Viii, light pink carbons) with N5 deprotonated and no methanol. DFT model of species V and VI are shown in d and e, respectively, and are overlaid in f (V green carbons, VI magenta carbons). g, Overlay of the species VI DFT model (magenta carbons) with the crystal coordinates (teal carbons). Models were geometry optimized in the gas phase using the B3LYP/6-311++G(d,p) level of theory. Harmonic vibrational frequencies calculated using normal mode analysis were used to confirm that optimized geometries of all species were in local or global minima. In the case of species Vi, ‘ModRedundant’ optimisation was performed to fix the C–N5 distance and one imaginary frequency of 67.60 cm−1 was observed. Cartesian coordinates of the optimized structures are given in the Supplementary Information.
Supplementary information
Supplementary Data 1
Cartesian coordinates of optimized DFT models. (DOC 54 kb)
Rights and permissions
About this article
Cite this article
White, M., Payne, K., Fisher, K. et al. UbiX is a flavin prenyltransferase required for bacterial ubiquinone biosynthesis. Nature 522, 502–506 (2015). https://doi.org/10.1038/nature14559
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14559
This article is cited by
-
In vitro construction of the COQ metabolon unveils the molecular determinants of coenzyme Q biosynthesis
Nature Catalysis (2024)
-
Isophthalate:coenzyme A ligase initiates anaerobic degradation of xenobiotic isophthalate
BMC Microbiology (2022)
-
Flavin-enabled reductive and oxidative epoxide ring opening reactions
Nature Communications (2022)
-
Critical enzyme reactions in aromatic catabolism for microbial lignin conversion
Nature Catalysis (2022)
-
An enzymatic activation of formaldehyde for nucleotide methylation
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.