Abstract
Understanding the diversity of human tissues is fundamental to disease and requires linking genetic information, which is identical in most of an individual’s cells, with epigenetic mechanisms that could have tissue-specific roles. Surveys of DNA methylation in human tissues have established a complex landscape including both tissue-specific and invariant methylation patterns1,2. Here we report high coverage methylomes that catalogue cytosine methylation in all contexts for the major human organ systems, integrated with matched transcriptomes and genomic sequence. By combining these diverse data types with each individuals’ phased genome3, we identified widespread tissue-specific differential CG methylation (mCG), partially methylated domains, allele-specific methylation and transcription, and the unexpected presence of non-CG methylation (mCH) in almost all human tissues. mCH correlated with tissue-specific functions, and using this mark, we made novel predictions of genes that escape X-chromosome inactivation in specific tissues. Overall, DNA methylation in several genomic contexts varies substantially among human tissues.
Similar content being viewed by others
Main
To understand the variability of DNA methylation across human tissues better, we obtained post-mortem samples of 18 tissue types from 4 individuals (5 singletons, 8 duplicates and 5 triplicates; Fig. 1a, Supplementary Methods and Supplementary Table 1) and performed deep transcriptome (36 messenger-RNA-seq samples; 120–475 million reads per sample), base-resolution methylome (36 MethylC-seq4 samples; 30–80× genome coverage per sample), and genome sequencing (4 whole genome sequences; 20–45× genome coverage per sample). We focused our initial analysis on cytosines in the CG context and used a previously published method2 to identify differential methylation (Supplementary Methods). We found that 15.4% (4,073,896 out of 26,474,560 sites tested) of CG sites in these experiments are strongly differentially methylated (minimum methylation difference ≥ 0.3; Extended Data Fig. 1a), which is similar to a previous study2. To identify differentially methylated regions (DMRs), we combined sites within 500 base pairs (bp) of one another and found 1,198,132 DMRs. Even with these stringent criteria, 719,837 (60.1%) of the DMRs we identified were novel2,5.
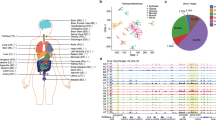
a, The tissues analysed in this study. Samples are denoted by the two letter code in parentheses followed by an individual ID. b, Browser screenshot of an example DMR. The top track contains gene models. The following four tracks contain green blocks indicating the location of super enhancers, enhancers and hypomethylated DMRs in the aorta, respectively. The remaining tracks display methylation data from each sample. Gold ticks are CG sites with heights proportional to their methylation level. Ticks on the forward and reverse strand are projected upward and downward from the dotted line, respectively. c, d, Hierarchical clustering of DMR methylation levels (c) and expression levels of differentially expressed genes (d). Colours indicate the organ systems each sample belongs to.
As expected, hypomethylation at DMRs correlated with tissue-specific functions2,6. For example, strongly hypomethylated DMRs in the aorta overlap with aorta-specific super enhancers7 around MYH10, a gene involved in blood vessel function8 (Fig. 1b). To validate our DMRs further, we performed hierarchical clustering on their weighted methylation levels9 (Supplementary Methods, Fig. 1c and Extended Data Fig. 1b, c). Tissues that were part of the same organ system clustered together (for example, heart and muscle tissues). We compared these results to a clustering of differentially expressed genes identified in the transcriptomes and found a similar separation of organ systems (Supplementary Methods, Fig. 1d and Extended Data Fig. 1d). Furthermore, Genomic Regions Enrichment of Annotations Tool10 analysis on the most hypomethylated tissue-specific DMRs revealed many tissue-specific functions (Extended Data Fig. 1e, f, Supplementary Methods and Supplementary Tables 2,3).
To examine the relationship between methylation and transcription, we correlated the methylation levels of DMRs and the expression of the closest genes (Fig. 2a, Extended Data Fig. 2a, b and Supplementary Methods). As expected, methylation in DMRs had a negative correlation with expression, and this correlation grew stronger closer to the transcription start site. The strongest negative correlation was not in gene promoters but downstream of the promoter up to 8 kilobases (kb) away (intragenic (0.3 kb to 8 kb) versus promoter region and upstream region (−2 kb to 0.3 kb) median Spearman correlation coefficient difference −0.07; Mann–Whitney P = 4.2 × 10−17; Fig. 2a). This analysis shows that transcription is strongly associated with intragenic DMRs in the tissues we examined, extending similar observations in cancer methylomes11.
a, The mean Spearman correlation coefficient at various distances between the methylation level of autosomal DMRs and the expression of the nearest gene. These correlations are shown for DMRs: overlapping genes (gene body), overlapping enhancers, overlapping promoters or CpG islands (CGIs) or CGI shores, not overlapping genes (intergenic) and all remaining DMRs (undefined). TSS, transcription start site. b, Heatmap showing the tissue-specific methylation preference of each motif. The tissues are coloured according to Fig. 1c, and the ordering is listed at the bottom of the figure. The bar plot on the right shows the number of times the motif was present in the 20 motif models. c, The number of base pairs covered by PMDs in all samples. d, The distribution of expression inside and outside of PA-2 PMDs across various samples. Notches indicate a confidence interval estimated from 1,000 bootstrap samples. Each PMD boxplot consists of 3,627 genes, and each non-PMD boxplot consists of 22,907 genes. FPKM, fragments per kilobase of transcript per million mapped reads. e, f, Histone modification profiles in and around PMDs in PA-2 (e) and IMR90 (f).
These intragenic methylation differences have previously been suggested to mark intragenic CG islands (CGIs) or CGI shores5,12,13,14. However, only a small fraction of intragenic DMRs fell in these features (19%; Extended Data Fig. 2c). In addition, predicted enhancers and putative promoters only accounted for 23% and 22% of intragenic DMRs, respectively, suggesting that the remaining DMRs, which we call undefined intragenic DMRs (uiDMRs), represent an unrecognized set of functional elements (35%; Extended Data Fig. 2c and Supplementary Methods). The methylation level of these uiDMRs correlated strongly with the expression of the genes containing them. To examine their regulatory potential, we plotted their histone modification profiles (histone 3 Lys 4 methylation (H3K4me1), H3K4me3, H3K27ac, H3K9me3, H3k27me3 and H3K36me3) derived from the same tissue samples15 and found five classes: weak enhancer, promoter-proximal, transcribed, poised enhancer and unmarked (Extended Data Figs 2d–h, 3a, b and Supplementary Methods). Classes with strong, active histone modifications were moderately negatively correlated with expression (weak enhancer and proximal promoter uiDMRs; median Spearman correlation coefficient −0.32 and −0.16, respectively); whereas, uiDMRs with less active histone modifications exhibited a weak negative correlation (transcribed and poised enhancer uiDMRs). Notably, the correlation between expression and methylation at promoter-proximal uiDMRs was as strong as the correlation with intragenic DMRs that overlapped strong promoters (Extended Data Fig. 4 and Supplementary Methods), indicating that intragenic promoter and promoter-proximal sequences are more predictive of changes in methylation than those enriched for enhancer-like chromatin modifications.
By contrast, unmarked uiDMRs showed a weakly positive correlation with expression (Extended Data Fig. 4d). Notably, we found many of the motifs enriched in tissue-specific uiDMRs were present in tissue-specific enhancers (for example, HNF4a (ref. 16) in liver-specific uiDMRs), suggesting that these DMRs are tissue-specific regulatory elements (Supplementary Methods and Supplementary Tables 4 and 5). Recently, hypomethylated regions that appear inactive in adult tissues but active during fetal development were identified in mice6. We examined the DNase I hypersensitivity profiles of unmarked uiDMRs in matched fetal tissues17 and found an enrichment of hypersensitivity (Extended Data Fig. 5 and Supplementary Table 6), suggesting that hypomethylation of inactive DMRs can be maintained at regions active earlier in development.
We next examined whether variation in methylation is associated with genetic variation across individuals, which has not been widely characterized in healthy primary tissues or using whole-genome bisulphite sequencing18,19.To identify individual-specific DMRs, we used a method20 that is sensitive to these differences unlike the methodology used above (Supplementary Methods). We first restricted our analysis to triplicated samples and ranked DMRs by a tissue-specific methylation outlier score that is largest when the methylation level in one individual differs from the other two. We found an ∼1.6-fold enrichment of single nucleotide polymorphisms (SNPs) associating with methylation changes in the top 2,500 methylation-outlier-score-ranked DMRs in all tissues (Supplementary Methods). We then used the Epigram pipeline21 to predict tissue-specific methylation from DNA motifs in these DMRs and found them highly predictive (average area under the curve (AUC) 0.79; Supplementary Methods). These full models used an average of 156 motifs; however, an average AUC of 0.74 was achieved using only 20 core transcription factor motifs per tissue.
We then identified groups of corresponding motifs by clustering the sets of tissue-specific motifs (Supplementary Methods). The motif groups were clustered by their tissue hypo- and hypermethylation specificities (Fig. 2b). In total, 42 out of 95 motifs only had hypomethylation specificity; for example, MEIS, which is involved in heart development22, is hypomethylated in the left ventricle, right atrium and right ventricle. We also identified 34 motifs enriched at both hypo-methylated DMRs in some tissues, and in hyper-methylated DMRs in some other tissues. Three of these motifs match transcription factor families (FOX, HOX and GATA) and are most significantly enriched in hypomethylated regions, suggesting that they are primarily involved in regulating hypomethylation.
Mammalian cells have high genome-wide levels of mCG, with the exception of a cultured human fetal fibroblast cell line (IMR90)4, cancer cells23,24 and placenta (PLA)25. Surprisingly, large regions of the pancreatic methylomes (PA-2 and PA-3) were significantly hypomethylated (Extended Data Fig. 6a). We developed a method to identify partially methylated domains (PMDs) genome-wide (Supplementary Tables 7,8 and Supplementary Methods) and found pancreatic PMDs were smaller than those in IMR90 and PLA (Extended Data Fig. 6b) and covered a smaller fraction of the genome (Fig. 2c). All pairs of PMDs overlapped significantly, indicating that these regions are largely shared (>40% overlap; P < 0.001; Extended Data Fig. 6c).
Genes in samples with PMDs are transcriptionally repressed25,26, but these regions also show reduced expression in all of the tissues we surveyed whether or not a PMD is present (Fig. 2d). In both IMR90 and PA-2, these regions showed an enrichment in repressive modifications (H3K27me3 and H3K9me3; median difference 0.025–0.168 reads per kilobase per million (RPKM); Mann–Whitney P < 2.51 × 10−161) and a depletion in active modifications (H3K4me1, H3K27ac and H3K36me3; median difference 0.050–0.012 RPKM; Mann–Whitney P < 2.03 × 10−53) compared to shuffled regions (Fig. 2e, f, Extended Data Fig. 6 d, e and Supplementary Methods), which provides a potential mechanism for their repression. To try to account for this global hypomethylation, we plotted the expression levels of DNMT1, DNMT3A, DNMT3B and DNMT3L but found no systematic expression difference between samples with and without PMDs (Extended Data Fig. 7a–d).
Previous studies have highlighted the existence of methylation outside of the CG context (mCH) in human embryonic stem cells4, brain1,20 and at the promoter of the PGC-1α gene (PPARGC1A) in skeletal muscle27. We found evidence for appreciable amounts of mCH in many of these tissues (Fig. 3a and Extended Data Fig. 8a). A 5-bp motif split the samples into two groups, one with mCH enriched in a TNCAC motif and another with mCH enriched in an NNCAN motif (where N is any base) (Supplementary Methods). The TNCAC motif is highly similar to the one previously identified in purified glia (GLA) and neurons (NRN) (TACAC). These motifs differ from those found in H1 embryonic stem cells (H1) and induced pluripotent stem cells (TACAG)4,26 (Fig. 3b–d). We quantified the extent of mCH across these samples by plotting the distribution of methylation levels at mCH sites in the 25 samples with a TNCAC motif, which revealed a methylation level similar to that of GLA, NRN and H1 (Extended Data Fig. 8b)4,20. Most of the tissue types were consistently enriched for the TNCAC or NNCAN motif, but several (oesophagus, lung, pancreas and spleen) had replicates that disagreed, suggesting that mCH is not homogenously distributed across these tissues.
a, The fraction of methylated cytosines in the CH context by sample. b–d, Representative mCH motifs from embryonic (H1; b), tissue (LI-11; c) and brain (NRN; d) samples. The height of each letter represents its information content. e, Heatmap of genic mCAS patterns normalized to the flanking region. Each gene was assigned to 1 of 20 clusters, which is indicated by the number and tick marks on the y axis. The tick marks on the x axis indicate the upstream, transcription start, transcription end, and downstream segments of each gene. The boxes around various patterns highlight regions referenced in the main text. TES, transcription end site. f, Bar plot of the ratio of the genome-wide mCAC to mCAG in various samples.
To examine the potential functional effect of mCH in adult tissues, we plotted the distribution of expression levels for various quantiles of gene body mCH as it was previously reported to be positively correlated with expression in H1 (ref. 4) and negatively correlated with expression in neurons20. This analysis revealed a negative correlation between expression and mCH (Extended Data Fig. 8c and Supplementary Methods). Next, we combined our replicates and clustered genes by the patterns of CAS methylation (in which S is a G or C) in and around their gene body (Fig. 3e and Supplementary Methods). To characterize the genes assigned to each cluster, we performed DAVID functional annotation clustering (Supplementary Table 9 and Supplementary Methods), which revealed several different classes. Clusters 1, 2, 16 and 19 contained genes highly enriched for terms involved in basic cellular processes and had an active methylation state (that is, hypermethylation in embryonic samples and hypomethylation in tissue and brain samples) across all samples. Clusters 5 and 6 were dominated by terms related to neuronal function and genes in this class were differentially methylated between neurons and glia and have inactive methylation states in other samples (that is, hypomethylation in embryonic samples and hypermethylation in tissue and brain samples). Cluster 12 was enriched for heart- and muscle-related terms and its genes had an active methylation state in the three heart tissues as well as a weakly active methylation state in psoas but appeared inactive in other samples. Lastly, cluster 14 possessed an active methylation state in brain and tissue samples but was inactive in embryonic samples. Despite being inactive in the H1 samples, this class of genes was highly enriched for terms related to development.
To define the transition of mCH motifs over development better, we examined the ratio of the methylation level of CAC and CAG (mCAC and mCAG) sites in a variety of differentiated (tissues, NRN and GLA), embryonic (H1), and embryonic-derived (neural progenitor cells (NPC), mesendoderm (MES), trophoblast-like (TRO), mesenchymal stem cells (MSC))28 cell samples (Fig. 3f). With the exception of brain cells, mCH levels drop during differentiation, and the mCAC/mCAG ratios revealed a shift in motif usage across developmental time (Fig. 3f); although, mCAC and mCAG within the same gene remain tightly correlated in both early embryonic and differentiated tissues (Extended Data Fig. 8d, e).
Methylation has previously been shown to be predictive of genes escaping X-chromosome inactivation in neurons20. We investigated this phenomenon in these samples by comparing the promoter mCG and gene body mCH of genes that had previously been identified to escape X-chromosome inactivation29 in 11 tissues with mCH (Fig. 4a). Female-specific promoter mCG hypomethylation and gene body mCH hypermethylation were present at escapee genes at a similar level as in neurons20 (Extended Data Fig. 9a). Using these tissue methylomes, gene body mCH was appreciably predictive of biallecially expressed genes (AUC 0.89; Extended Data Fig. 9b and Supplementary Methods). To a lesser extent, we observed female-specific promoter mCH and gene body mCG hypermethylation at escapee genes (Extended Data Fig. 9a, c, d). Although female-specific promoter mCG hypomethylation, promoter mCH hypermethylation and gene body mCG hypermethylation are predictive of X-chromosome inactivation escapees, female-specific gene body mCH hypermethylation is the most predictive feature of X-chromosome inactivation escapees (Extended Data Fig. 9a, b–e). We detected female-specific mCH hypermethylation in 109 out of 612 X-linked genes, including 9 genes hypermethylated in all 11 tissues and 72 genes that were hypermethylated in only one tissue (Fig. 4b). Several genes such as FUNDC1 showed female-specific hypermethylation in several tissues but not in neurons, suggesting a tissue-dependent regulation of the escape from X inactivation.
a, Browser screenshot of the increase in female mCH for a gene known to escape X-chromosome inactivation (MED14). Sample names are coloured by gender (male, black; female, red). MED14-AS1 is also known as MED14OS. b, Ratio of mCH level in female versus male samples across genes with a significant difference in at least one sample. Cells boxed in black denote samples with a statistically significant difference between females and males. The XCI score for each gene is from ref. 29 and indicates the degree of escaping X-chromosome inactivation. c, The number of ASM and ASE sites across the triplicated tissues. The top row depicts ASM events (left) and ASE events (right) that are allele-specific in all tissues (black), are variable across tissues (white), or do not possess enough data to tell (grey). The bottom row depicts the distribution of variable sites from the top row that vary by individual (blue), tissue (red) or neither (purple).
Allele-specific methylation and expression (ASM and ASE, respectively) may also have a role in the regulation of autosomal genes. To examine these phenomena in human tissues, we combined the RNA-seq and MethylC-seq data sets with phased genotypes for each individual in this study3,15 (Extended Data Fig. 10a and Supplementary Methods). Using the triplicate tissue samples (fat (FT), gastric (GA), psoas (PO), small bowel (SB) and spleen (SX)), we identified 8,464–48,560 ASM events in the CG context and 48–403 ASE genes across these tissues (Supplementary Tables 10, 11 and Supplementary Methods). We next looked for ASM events that varied across individuals within a tissue-type (tissue variable) and those that varied across a tissue-type within an individual (individual variable). Of the ASM events that varied, 4.1–7.5% and 54.5–70.0% were tissue- and individual-variable, respectively; whereas, of the ASE events that varied, 0.0–20.0% were individual-variable and 13.3–48.8% were tissue-variable (Fig. 4c and Supplementary Methods). Of the ASE events, 38.4–87.4% had an ASM event within 100 kb, and of these sites, 76% had an ASM and ASE event that was matched (that is, a DMR was hypomethylated on the same haplotype as the more highly expressed allele). Furthermore, we found that a larger fraction of ASE genes were observed near ASM events whether or not the events matched (Extended Data Fig. 10 b, c and Supplementary Methods). These results demonstrate a link between ASM and ASE in human tissues.
Here we have presented the deepest set of base resolution maps of mCG and mCH so far along with chromatin modification states, haplotype-resolved genome sequences and transcriptional profiles for a large set of human tissues. These data sets allowed us to identify cis-regulatory elements accurately. Furthermore, they revealed the existence of mCH genome-wide in a subpopulation of cells from differentiated human tissues, which seems to be repressive. Our analysis of genic mCH across human tissues indicates a tissue-specific distribution that is distinct from those genes that were previously identified in embryonic stem cells and the brain. These genes are enriched for a variety of functions, most surprisingly those involved in development. These analyses raise the intriguing possibility that mCH is used in adult stem cells30 and could help to repress these genes as the cells transition into their differentiated role.
Accession codes
Primary accessions
Gene Expression Omnibus
Sequence Read Archive
Data deposits
The sequencing data sets generated for this study as well as those for the IMR90, H1 and H1 derived samples can be found at the Gene Expression Omnibus (GEO) under the accession number GSE16256. The sequencing data sets for the fetal tissues used in this study can be found at GEO under the accession number GSE18927. The sequencing data sets for the placental tissue used in this study can be found at GEO under the accession number GSE39777. The sequencing data sets for the neuronal and glial samples can be found at GEO under the accession number GSE47966 (NRN GSM1173776; GLA GSM1173777). The human tissue sequencing data generated for this study can be found at Sequence Read Archive (SRA) under the project number SRP000941. Analysed data sets can be obtained from http://neomorph.salk.edu/human_tissue_methylomes.html.
References
Varley, K. E. et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 23, 555–567 (2013).
Ziller, M. J. et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 500, 477–481 (2013).
Selvaraj, S., Dixon, J. R., Bansal, V. & Ren, B. Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nature Biotechnol. 31, 1111–1118 (2013).
Lister, R. et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322 (2009).
Irizarry, R. A. et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nature Genet. 41, 178–186 (2009).
Hon, G. C. et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nature Genet. 45, 1198–1206 (2013).
Hnisz, D. et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013).
Yuen, S. L., Ogut, O. & Brozovich, F. V. Nonmuscle myosin is regulated during smooth muscle contraction. Am. J. Physiol. Heart Circ. Physiol. 297, H191–H199 (2009).
Schultz, M. D., Schmitz, R. J. & Ecker, J. R. ‘Leveling’ the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 28, 583–585 (2012).
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nature Biotechnol. 28, 495–501 (2010).
Hovestadt, V. et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 510, 537–541 (2014).
Maunakea, A. K. et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466, 253–257 (2010).
Doi, A. et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nature Genet. 41, 1350–1353 (2009).
Deaton, A. M. et al. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 21, 1074–1086 (2011).
Leung, D. et al. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature 518, 350–354 (2015).
Parviz, F. et al. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nature Genet. 34, 292–296 (2003).
Maurano, M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Gutierrez-Arcelus, M. et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife 2, e00523 (2013).
Liu, Y. et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nature Biotechnol. 31, 142–147 (2013).
Lister, R. et al. Global epigenomic reconfiguration during mammalian brain development. Science 341, 6146 (2013).
Whitaker, J. W., Chen, Z. & Wang, W. Predicting the human epigenome from DNA motifs. Nature Methods 12, 265–272 (2015).
Stankunas, K. et al. Pbx/Meis deficiencies demonstrate multigenetic origins of congenital heart disease. Circ. Res. 103, 702–709 (2008).
Hon, G. C. et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 22, 246–258 (2012).
Berman, B. P. et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nature Genet. 44, 40–46 (2011).
Schroeder, D. I. et al. The human placenta methylome. Proc. Natl Acad. Sci. USA 110, 6037–6042 (2013).
Lister, R. et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68–73 (2011).
Barrès, R. et al. Non-CpG methylation of the PGC-1α promoter through DNMT3B controls mitochondrial density. Cell Metab. 10, 189–198 (2009).
Xie, W. et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153, 1134–1148 (2013).
Carrel, L. & Willard, H. F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434, 400–404 (2005).
Wagers, A. J. & Weissman, I. L. Plasticity of adult stem cells. Cell 116, 639–648 (2004).
Acknowledgements
We thank R. J. Schmitz for critical reading of the manuscript. This work is supported by the National Institutes of Health (NIH) Epigenome Roadmap Project (U01 ES017166). E.A.M. was supported by National Institute of Neurological Diseases and Stroke grant (R00NS080911). J.R.E. was supported by the Gordon and Betty Moore Foundation (GMBF3034) and the Mary K. Chapman Foundation. T.J.S. and J.R.E. are investigators of the Howard Hughes Medical Institute. S.L. was supported by NIH fellowship grants F32HL110473 and K99HL119617. The authors acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing HPC resources that have contributed to the research results reported within this paper. The authors would also like to thank Mid-America Transplant Services, St Louis, for their support of this research effort.
Author information
Authors and Affiliations
Contributions
B.R., T.J.S., W.W. and J.R.E. designed and supervised research. S.L. and Y.L. collected tissues. J.R.N. and M.A.U. conducted MethylC-seq, RNA-seq and genome sequencing experiments. D.L. conducted ChIP-seq experiments. N.R. performed ChIP-seq data analysis. M.D.S., Y.H., M.H. and H.C. performed sequencing data processing. J.W.W. performed motif prediction and mutation analysis. M.D.S. designed and implemented the methylation processing and analysis module. M.D.S., Y.H., J.W.W., M.H. and E.A.M. performed statistical and bioinformatic analyses. M.D.S., Y.H., J.W.W. and J.R.E. prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Identification of DMRs and multidimensional scaling analysis.
a, Line plot showing the fraction of differentially methylated CG sites (dynamic CGs) out of all CG sites under various methylation difference cutoffs. The methylation difference of a CG site is defined in ref. 2. The grey line indicates the cutoff (0.3) used to call differentially methylated sites. b, A plot of the first two principal components from the methylation level multidimensional scaling. Tissues are shaded by the organ group they belong to as in Fig. 1c, d. c, d, Bar charts of the cumulative amount of variance explained by the first N principal components from the multidimensional scaling performed on the methylation levels of all DMRs (c) and the expression levels of all differentially expressed genes (d). e, A representative example of enriched Gene Ontology biological process terms based on the most hypomethylated DMRs from LV-1. f, A representative example of enriched mouse phenotype terms based on the most hypomethylated DMRs from LV-1.
Extended Data Figure 2 DMRs and their correlation with transcription.
a, A browser screenshot of an example DMR downstream of the transcription start site. b, Expression level of the BIN1 gene that contains the DMR in a. c, The percentages of hypomethylated intragenic DMRs in each class of genomic features. d–h, Histone modification profiles of five categories of uiDMRs.
Extended Data Figure 3 Classification of uiDMR histone profiles and uiDMR properties.
a, Heatmap of the histone modification profiles for the five types of uiDMRs. The profiles were plotted for each mark across the DMR and the 5 kb upstream and downstream and the colours of each cell indicate the input normalized ChIP-seq RPKM. The colours on the left indicate the group of each profile assigned by k-means clustering (red, weak enhancer; orange, promoter-proximal; green, transcribed; blue, unmarked; black poised enhancer). b, A pie chart of the distribution of uiDMRs across the classes defined by k-means clustering.
Extended Data Figure 4 Classification of promoter histone profiles.
a, A heatmap of the histone modification profiles across strong (rows labelled with red) and unmarked (rows labelled with orange) promoters. The profiles were plotted for each mark across the promoter and the 5-kb upstream and downstream, and the colours of each cell indicate the input normalized ChIP-seq RPKM. b, c, The aggregate profiles for strong (b) and unmarked (c) promoters, respectively. d, The distribution of the Spearman correlation coefficients between the methylation level of different types of hypomethylated intragenic DMRs and the expression of the nearest gene. Notches indicate a confidence interval estimated from 1,000 bootstrap samples.
Extended Data Figure 5 uiDMR fetal DNase I profiles.
DNase I profiles of various fetal tissues corresponding to the tissues presented in this study. The samples are arranged column-wise by age, and row-wise by fetal tissue. The uiDMR-unmarked line represents the DNase I profile of uiDMRs without histone modifications. The DMR-enhancer line represents the DNase I profile of DMRs that overlapped an enhancer in a matched tissue in this study (indicated in the row label in parentheses). The shuffled line represents the DNase I profile of uiDMRs randomly shuffled across the genome.
Extended Data Figure 6 PMD features.
a, A browser screenshot (see Fig. 1 for description) of an example PMD found in IMR90, PLA, PA-2 and PA-3. RV-1 is included as a representative sample without PMDs. b, The distribution of sizes of PMDs in various samples. c, A heatmap representation of the overlap between various sets of PMDs. The denominator of the fraction of overlap is determined by the sample on the y axis. d, e, ChIP-seq profiles of the PMD regions defined in PA-2 (c) and IMR90 (d) after shuffling.
Extended Data Figure 7 DNMT expression across tissues.
a–d, Bar plots of the expression (measured in log10 FPKMs) of DNMT1 (a), DNMT3A (b), DNMT3B (c) and DNMT3L (d) across various samples.
Extended Data Figure 8 mCH distribution and correlation.
a, A browser screenshot (see Fig. 1 for description) of an example region with non-CG methylation (mCH). Purple and pink ticks are methylated CHG and CHH sites, respectively (H = A, C or T). Ticks on the forward strand are projected upward from the dotted line and ticks on the reverse strand are projected downward. b, The distribution of methylation levels at mCH sites across all samples with a discernible TNCAC motif. Only mCH sites with at least 10 reads and a significant amount of methylation were considered. c, Boxplots of the expression values across different quantiles of CAC gene body methylation (gene body mCAC). d, Scatterplot of mCAG versus mCAC inside gene bodies. e, Bar plot of the correlation of mCAG and mCAC inside gene bodies (blue) and the theoretical maximal correlation (red) if mCAC and mCAG are perfectly correlated. f–h, The methylation levels of C (top), CG (middle) and CH (bottom) across the read positions for PO-2 (red line) and EG-3 (blue line). Vertical lines indicate the position (tenth base from the beginning) where trimming was applied. i, mCH motif from PO-2 with the first 10 bases of each read trimmed. j, mCH motif from PO-2 without trimming. k, mCH motif from EG-3 with the first 10 bases of each read trimmed. l, mCH motif from EG-3 without trimming. The height of each letter represents its information content (that is, prevalence).
Extended Data Figure 9 X-chromosome inactivation.
a, Distributions of promoter CG methylation (mCG) levels (mCG/CG), gene body non-CG methylation (mCH) levels (mCH/CH), gene body mCG levels and promoter mCH levels in genes previously reported to express from only one allele (inactivated) or biallelically (escapee)29. Black ticks show median, and bars indicate the twenty-fifth to seventy-fifth percentile range. Genes more prone to escaping inactivation have lower promoter mCG, higher gene body mCH, higher gene body mCG and higher promoter mCH in females. b–e, Discriminability analysis using gender-specific gene body mCH (b), promoter mCG (c), gene body mCG (d) and promoter mCH (e) to predict the escapee status of X-linked genes, respectively. Among them, gene body mCH is the most predictive feature of X-chromosome inactivation escapees. The discriminability was measured by the area under the curve (AUC) (Supplementary Methods).
Extended Data Figure 10 ASMand ASE.
a, An example of ASM. Reads that contain a heterozygous SNP (red box) are separated by allele. The number of methylated (reads containing Cs) and unmethylated (reads containing Ts) at adjacent CG sites (black boxes) are tested for differential methylation. b, Fraction of ASE genes (blue) and bi-allelically expressed genes (grey) that have at least one ASM event within a certain distance. Bi-allelically expressed genes were defined as genes that were covered by at least 10 reads and whose P values, given by binomial test for allelic expression, were greater than 0.2 (that is, no significance). c, Fraction of ASE genes that were linked to matched ASM events (blue) and matched ASM events with their locations shuffled (grey). b, c, Aggregated results using samples from triplicate tissues.
Supplementary information
Supplementary Information
This file contains Supplementary Methods and additional references. (PDF 976 kb)
Supplementary Tables
This zipped file contains Supplementary Tables 1-11. (ZIP 18279 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported licence. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons licence, users will need to obtain permission from the licence holder to reproduce the material. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-sa/3.0/.
About this article
Cite this article
Schultz, M., He, Y., Whitaker, J. et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 523, 212–216 (2015). https://doi.org/10.1038/nature14465
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14465
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.