Abstract
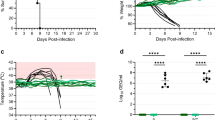
The family Filoviridae contains three genera, Ebolavirus (EBOV), Marburg virus, and Cuevavirus1. Some members of the EBOV genus, including Zaire ebolavirus (ZEBOV), can cause lethal haemorrhagic fever in humans. During 2014 an unprecedented ZEBOV outbreak occurred in West Africa and is still ongoing, resulting in over 10,000 deaths, and causing global concern of uncontrolled disease. To meet this challenge a rapid-acting vaccine is needed. Many vaccine approaches have shown promise in being able to protect nonhuman primates against ZEBOV2. In response to the current ZEBOV outbreak several of these vaccines have been fast tracked for human use. However, it is not known whether any of these vaccines can provide protection against the new outbreak Makona strain of ZEBOV. One of these approaches is a first-generation recombinant vesicular stomatitis virus (rVSV)-based vaccine expressing the ZEBOV glycoprotein (GP) (rVSV/ZEBOV). To address safety concerns associated with this vector, we developed two candidate, further-attenuated rVSV/ZEBOV vaccines. Both attenuated vaccines produced an approximately tenfold lower vaccine-associated viraemia compared to the first-generation vaccine and both provided complete, single-dose protection of macaques from lethal challenge with the Makona outbreak strain of ZEBOV.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Feldman, H., Sanchez, A. & Geisbert, T. W. in Fields Virology Ch. 32 (Lippincott Williams and Wilkins, 2013).
Mire, C. E. & Geisbert, T. W. in Biology and Pathology of Rhabdo- and Filoviruses Ch. 22 (World Scientific Publishing, 2014).
Leroy, E. M. et al. Fruit bats as reservoirs of Ebola virus. Nature 438 575–576 (2005).
Stanley, D. A. et al. Chimpanzee adenovirus vaccine generates acute and durable protective immunity against ebolavirus challenge. Nature Med. 20 1126–1129 (2014).
Volchkova, V. A., Dolnik, O., Martinez, M. J., Reynard, O. & Volchkov, V. E. Genomic RNA editing and its impact on Ebola virus adaptation during serial passages in cell culture and infection of guinea pigs. J. Infect. Dis. 204 (Suppl 3). S941–S946 (2011).
Kugelman, J. R. et al. Ebola virus genome plasticity as a marker of its passaging history: a comparison of in vitro passaging to non-human primate infection. PLoS ONE 7 e50316 (2012).
Mohan, G. S., Li, W., Ye, L., Compans, R. W. & Yang, C. Antigenic subversion: a novel mechanism of host immune evasion by Ebola virus. PLoS Pathog. 8 e1003065 (2012).
Hirschberg, R. et al. Challenges, progress, and opportunities: proceedings of the filovirus medical countermeasures workshop. Viruses 6 2673–2697 (2014).
Jones, S. M. et al. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nature Med. 11 786–790 (2005).
Feldmann, H. et al. Effective post-exposure treatment of Ebola infection. PLoS Pathog. 3 e2 (2007).
Mire, C. E. et al. Recombinant vesicular stomatitis virus vaccine vectors expressing filovirus glycoproteins lack neurovirulence in nonhuman primates. PLoS Negl. Trop. Dis. 6, e1567 (2012).
Clarke, D. K. et al. Synergistic attenuation of vesicular stomatitis virus by combination of specific G gene truncations and N gene translocations. J. Virol. 81 2056–2064 (2007).
Clarke, D. K. et al. Neurovirulence and immunogenicity of attenuated recombinant vesicular stomatitis viruses in nonhuman primates. J. Virol. 88 6690–6701 (2014).
Johnson, J. E. et al. In vivo biodistribution of a highly attenuated recombinant vesicular stomatitis virus expressing HIV-1 Gag following intramuscular, intranasal, or intravenous inoculation. Vaccine 27 2930–2939 (2009).
Kugelman, J. R. et al. Evaluation of the potential impact of Ebola virus genomic drift on the efficacy of sequence-based candidate therapeutics. MBio 6 e02227–14 (2015).
Baize, S. et al. Emergence of Zaire Ebola virus disease in guinea. N. Engl. J. Med. 371 1418–1425 (2014).
Gire, S. K. et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science 345 1369–1372 (2014).
Hoenen, T. et al. Mutation rate and genotype variation of Ebola virus from Mali case sequences. Science http://dx.doi.org/10.1126/science.aaa5646 (2015).
Cooper, D. et al. Attenuation of recombinant vesicular stomatitis virus HIV-1 vaccine vectors by gene translocations and G gene truncation reduces neurovirulence and enhances immunogenicity in mice. J. Virol. 82 207–219 (2008).
Witko, S. E. et al. An efficient helper-virus-free method for rescue of recombinant paramyxoviruses and rhadoviruses from a cell line suitable for vaccine development. J. Virol. Methods 135 91–101 (2006).
Thi, E. P. et al. Marburg virus infection in nonhuman primates: Therapeutic treatment by lipid-encapsulated siRNA. Sci. Transl. Med. 6, 250ra116 (2014).
Acknowledgements
We thank V. Borisevich and D. Deer for assistance with clinical pathology assays performed in the GNL BSL-4 laboratory, and the staff of the UTMB Animal Resources Center for animal care. This study was supported by the funds from the Department of Microbiology and Immunology at UTMB to T.W.G. and in part by the Department of Health and Human Services, National Institutes of Health grant R01AI09881701 to J.H.E. and T.W.G.
Author information
Authors and Affiliations
Contributions
D.K.C., D.M., and T.E.L. designed the vaccine vectors and did preparative work. J.H.E., M.A.E., A.O.-S., and R.X. designed, conducted, and analysed the in vitro vaccine characterization studies. C.E.M., J.H.E., and T.W.G. conceived and designed the NHP study. C.E.M., J.B.G., and T.W.G. performed the NHP vaccination and challenge experiments, and conducted clinical observations of the animals. J.B.G. and K.N.A. performed the clinical pathology assays. J.B.G. performed the ZEBOV infectivity assays. C.E.M., D.M., J.B.G., K.N.A., M.A.E., K.A.F., D.K.C, J.H.E, and T.W.G. analysed the data. K.A.F. performed histologic and immunohistochemical analysis of the data. C.E.M., D.M., D.K.C., and T.W.G. wrote the paper. All authors had access to all of the data and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The N1 and N4 rVSV vectors described in this manuscript are the subject of patents licensed to Profectus BioSciences, Inc. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the University of Texas Medical Branch.
Extended data figures and tables
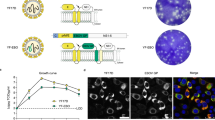
Extended Data Figure 1 Relative immunogenicity of rVSV/ZEBOV vectors in cynomolgus macaques.
At study day −28, cynomolgus macaques were immunized IM with 2 × 107 PFU of either N4 or N1 vectors. Ten days after a single immunization, PBMCs were prepared and ZEBOV GP-specific T-cell responses were quantitated by IFN-γ ELISpot assay. a, ZEBOV GP-specific IFN-γ ELISpot responses in individual macaques. b, Average ZEBOV GP-specific IFN-γ ELISpot responses with standard error of the means indicated.
Rights and permissions
About this article
Cite this article
Mire, C., Matassov, D., Geisbert, J. et al. Single-dose attenuated Vesiculovax vaccines protect primates against Ebola Makona virus. Nature 520, 688–691 (2015). https://doi.org/10.1038/nature14428
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14428
This article is cited by
-
Natural history of nonhuman primates after conjunctival exposure to Ebola virus
Scientific Reports (2023)
-
Recombinant MVA-prime elicits neutralizing antibody responses by inducing antigen-specific B cells in the germinal center
npj Vaccines (2021)
-
Prior vaccination with rVSV-ZEBOV does not interfere with but improves efficacy of postexposure antibody treatment
Nature Communications (2020)
-
A versatile platform technology for recombinant vaccines using non-propagative human parainfluenza virus type 2 vector
Scientific Reports (2019)
-
Vesicular Stomatitis Virus-Based Vaccine Protects Mice against Crimean-Congo Hemorrhagic Fever
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.