Abstract
Drug resistance invariably limits the clinical efficacy of targeted therapy with kinase inhibitors against cancer1,2. Here we show that targeted therapy with BRAF, ALK or EGFR kinase inhibitors induces a complex network of secreted signals in drug-stressed human and mouse melanoma and human lung adenocarcinoma cells. This therapy-induced secretome stimulates the outgrowth, dissemination and metastasis of drug-resistant cancer cell clones and supports the survival of drug-sensitive cancer cells, contributing to incomplete tumour regression. The tumour-promoting secretome of melanoma cells treated with the kinase inhibitor vemurafenib is driven by downregulation of the transcription factor FRA1. In situ transcriptome analysis of drug-resistant melanoma cells responding to the regressing tumour microenvironment revealed hyperactivation of several signalling pathways, most prominently the AKT pathway. Dual inhibition of RAF and the PI(3)K/AKT/mTOR intracellular signalling pathways blunted the outgrowth of the drug-resistant cell population in BRAF mutant human melanoma, suggesting this combination therapy as a strategy against tumour relapse. Thus, therapeutic inhibition of oncogenic drivers induces vast secretome changes in drug-sensitive cancer cells, paradoxically establishing a tumour microenvironment that supports the expansion of drug-resistant clones, but is susceptible to combination therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Engelman, J. A. & Settleman, J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr. Opin. Genet. Dev. 18, 73–79 (2008)
Holohan, C., Van Schaeybroeck, S., Longley, D. B. & Johnston, P. G. Cancer drug resistance: an evolving paradigm. Nature Rev. Cancer 13, 714–726 (2013)
Chapman, P. B. et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 364, 2507–2516 (2011)
Sosman, J. A. et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N. Engl. J. Med. 366, 707–714 (2012)
Shaw, A. T. & Engelman, J. A. ALK in lung cancer: past, present, and future. J. Clin. Oncol. 31, 1105–1111 (2013)
Zhou, C. et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 12, 735–742 (2011)
Villanueva, J. et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell 18, 683–695 (2010)
Nazarian, R. et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 468, 973–977 (2010)
Poulikakos, P. I. et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 480, 387–390 (2011)
Wagle, N. et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J. Clin. Oncol. 29, 3085–3096 (2011)
Lito, P. et al. Relief of profound feedback inhibition of mitogenic signaling by RAF inhibitors attenuates their activity in BRAFV600E melanomas. Cancer Cell 22, 668–682 (2012)
Shi, H. et al. Melanoma whole-exome sequencing identifies V600EB-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nature Commun. 3, 724–728 (2012)
Wilson, T. R. et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 487, 505–509 (2012)
Straussman, R. et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 487, 500–504 (2012)
Johannessen, C. M. et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature 504, 138–142 (2013)
Shi, H. et al. A novel AKT1 mutant amplifies an adaptive melanoma response to BRAF inhibition. Cancer Discovery 4, 69–79 (2014)
Diaz, L. A., Jr et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486, 537–540 (2012)
Shi, H. et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discovery 4, 80–93 (2014)
Sharma, S. V. et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80 (2010)
Kim, M. Y. et al. Tumor self-seeding by circulating cancer cells. Cell 139, 1315–1326 (2009)
Villanueva, J. & Herlyn, M. Melanoma and the tumor microenvironment. Curr. Oncol. Rep. 10, 439–446 (2008)
Acharyya, S. et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 150, 165–178 (2012)
Sun, Y. et al. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nature Med. 18, 1359–1368 (2012)
Lee, H. J. et al. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer Cell 26, 207–221 (2014)
Lito, P., Rosen, N. & Solit, D. B. Tumor adaptation and resistance to RAF inhibitors. Nature Med. 19, 1401–1409 (2013)
Sun, C. et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 508, 118–122 (2014)
Joseph, E. W. et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc. Natl Acad. Sci. USA 107, 14903–14908 (2010)
Heiman, M. et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell 135, 738–748 (2008)
Fuchs, Y. & Steller, H. Programmed cell death in animal development and disease. Cell 147, 742–758 (2011)
Kurtova, A. V. et al. Blocking PGE2-induced tumour repopulation abrogates bladder cancer chemoresistance. Nature 517, 209–213 (2015)
Tavazoie, S. F. et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451, 147–152 (2008)
Doyle, J. P. et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135, 749–762 (2008)
Zhang, X. H. et al. Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell 154, 1060–1073 (2013)
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013)
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010)
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57 (2009)
Supek, F., Bosnjak, M., Skunca, N. & Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800 (2011)
Valiente, M. et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell 156, 1002–1016 (2014)
Acknowledgements
We thank members of the Massagué laboratory for discussions; L. Sevenich and L. Akkari for technical advice. This work was supported by grants from the AACR (SU2C) to R.S.L., the MSK Metastasis Research Center, the NIH (CA163167 and CA129243), the Congressionally Directed Medical Research Program of the Department of Defense, the Howard Hughes Medical Institute, and the Cancer Center Support Grant P30 CA008748 to J.M., A.C.O. was an Erwin Schroedinger Fellowship awardee (J3013, FWF, Austrian Science Fund). A.L.J. was a Medical Research Fellow of the Howard Hughes Medical Institute. S.V. is supported by the Medical Research Council.
Author information
Authors and Affiliations
Contributions
A.C.O. and J.M. conceived the project, designed the experiments and wrote the paper. A.C.O. performed experiments and computational analysis. A.L.J., Y.Z., W.S. and T.W. assisted with experiments. Y.Z. and S.V. performed computational analysis. M.C.B. provided cell lines. X.K., H.S. and R.S.L. provided patient samples. N.R. provided clinical expertise, cell lines and drugs. All authors interpreted data, discussed results, and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Targeted therapy or oncogene knockdown leads to regression of sensitive melanoma and lung adenocarcinoma tumours but accelerates the proliferation and seeding of residual drug-resistant cells in vivo.
a, FACS analysis of sensitive A375 and vemurafenib-resistant A375R cells expressing TGL, at tumour implantation and after 2 weeks at start of therapy (n = 8 tumours). Plots depict representative images. b, Tumour volume of A375 cells treated with vehicle or vemurafenib over time (vehicle, n = 8; vemurafenib, n = 12 tumours). c, Representative sections of A375/A375R-TGL tumours at 0, 1, 3 and 6 days of vemurafenib treatment analysed with immunofluorescence against GFP. Arrowheads indicate emerging clusters of GFP+ resistant cells. Scale bars, 2 mm. d, Quantification of BrdU incorporation into vemurafenib-resistant A375R-TGL cells in A375/A375R tumours treated with vehicle or vemurafenib for 6 days (vehicle, n = 13 FOV of 3 tumours; vemurafenib n = 18 FOV of 4 tumours). Original magnification, ×20. e, Fold change of photon flux of TGL-expressing A375R cells in A375 tumours or A375R tumours alone treated with vehicle or dabrafenib for 8 days (A375/A375R: vehicle, n = 15; dabrafenib, n = 14; A375R: vehicle, n = 8; dabrafenib, n = 7 tumours). f, Tumour volume of doxycycline-inducible BRAF knockdown A375-i-shBRAF-derived xenograft tumours (in which ‘A375-i’ denotes expression of doxycycline-inducible hairpin) treated with vehicle or doxycycline over time (vehicle, n = 5; doxycycline, n = 4 tumours). g, Photon flux of TGL-expressing A375R cells mixed in A375-i-shBRAF tumours treated with vehicle or doxycycline (vehicle, n = 10; doxycycline, n = 11 tumours). h, Fold change of photon flux of TGL-expressing vemurafenib-resistant M249R4 tumours treated with vehicle or vemurafenib (n = 16 tumours). i–k, Co-implantation assay of tumours treated with vehicle or corresponding targeted therapy with BLI quantification after 5–8 days. i, Fold change of photon flux of TGL-expressing vemurafenib-resistant YUMM1.7R cells mixed in unlabelled, vemurafenib-sensitive YUMM1.7 tumours or YUMM1.7R tumours alone (YUMM1.7/YUMM1.7R: n = 24 ; YUMM1.7R: n = 20 tumours). j, Fold change of photon flux of TGL-expressing, intrinsically vemurafenib-resistant B16 cells mixed in vemurafenib-sensitive YUMM1.1 tumours or B16 tumours alone (YUMM1.1/B16: vehicle, n = 12; vemurafenib, n = 16; B16: n = 20 tumours). k, A375R mixed in crizotinib-sensitive H3122 cells or A375R tumours alone (H3122/A375R: vehicle, n = 14; crizotinib, n = 13; A375R: n = 12 tumours). l, Photon flux of tumours established from intrinsically resistant drug-resistant cells alone, treated with vehicle, crizotinib or erlotinib (crizotinib-resistant PC9, H2030- or erlotinib-resistant A375R) (n (from left to right) = 12, 12, 7, 12, 16 and 16 tumours, respectively). m, Summary of the model systems and conditions used in vivo. n, Left, representative immunofluorescence images of vemurafenib-treated, sensitive tumours 7 h or 5 days after intracardiac injection with A375R-TGL cells; sections stained for GFP (A375R, green), collagen type IV (blood vessels, red), and DAPI (nuclei, blue). Right, quantification of A375R single cells and cell clusters (≥2 cells) infiltrating an A375 tumour treated with vehicle or vemurafenib after intracardiac injection of A375R cells (GFP+ cells were scored in at least 10 whole sections of at least 4 tumours). Original magnifications, ×20. Data in b, e–l and n are mean and s.e.m., in f, centre line is median, whiskers are minimum to maximum. P values calculated by a two-tailed Mann–Whitney U test.
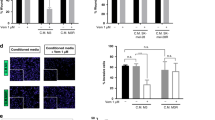
Extended Data Figure 2 The secretome of vemurafenib-treated melanoma and crizotinib- or erlotinib-treated lung adenocarcinoma cells stimulates the proliferation and migration of drug-resistant cells in vitro and occurs before apoptosis and senescence.
a, Quantification of the co-culture assay, depicted in Fig. 2a, 7 days after addition of resistant A375R-TGL cells (n = 4 biological replicates). P values calculated using a Student’s t-test. b, c, Drug-sensitive cells were pre-treated with vehicle or drug (crizotinib or erlotinib) for 48 h before 5 × 102 TGL-expressing, drug-resistant cells were added. Growth was monitored by BLI and quantified 7 days after addition of the resistant cell population (n = 8 biological replicates), P values calculated using a Student’s t-test. b, Quantification and representative images of TGL-expressing H2030 cells alone or co-cultured with crizotinib-sensitive H3122 cells and treated with vehicle or crizotinib c, Quantification and representative images of TGL-expressing A375R cells alone or co-cultured with erlotinib-sensitive HCC827 cells and treated with vehicle or erlotinib. d, Relative number of vemurafenib-resistant LOXR cells after 3 days in the presence of conditioned media derived from A375 and UACC62 cells (n = 3 biological replicates). e, Representative immunofluorescence for Ki67 in drug-resistant YUMM1.7R cells cultured in conditioned media from YUMM1.7 cells. Original magnification, ×20. f, Relative number of vemurafenib-resistant melanoma cells with different, clinically relevant resistance mechanisms after 3 days in the presence of conditioned media derived from A375 cells. SKMEL239-3 expressing the p61 BRAFV600E splice variant, A375 expressing NRASQ61K or the constitutively active MEK variant MEK-DD (n = 5 biological replicates). g, Relative cell number of intrinsically vemurafenib-resistant lung adenocarcioma cells (H2030, PC9) or crizotinib- and erlotinib-resistant melanoma cells (A375R) after 3 days cultured in the presence of conditioned media from vemurafenib-treated melanoma or crizotinib- and erlotinib-treated lung adenocarcinoma (n = 6 in all, except for A375R with HCC827-CM, n = 4 biological replicates). h, Representative image of A375R cells migrated towards A375-derived CM-vehicle or CM-vemurafenib. Original magnification, ×10. i, Relative migration towards conditioned media from different sources and different resistant test cells as indicated (n = 10 FOV). P < 0.01, P < 0.0001, two-tailed Mann–Whitney U test. j, Representative graph and quantification of real-time migration of A375R cells in the presence of conditioned media derived from A375 cells as measured by the xCELLigence system (n = 4 biological replicates). P value calculated using two-tailed Mann–Whitney U test. k, Monolayer gap closing assay of A375R cells in the presence of conditioned media derived from A375 cells with representative light microscopy images and quantification of gap closure over time. l, Immunoblotting for cleaved caspase-3 and phosphorylated ERK protein levels in vemurafenib-sensitive melanoma cell lines after 72 h of vemurafenib treatment. m, β-galactosidase staining of A375 cells treated with vemurafenib for 72 h or 8 days. Original magnification, ×13. Data are presented mean and s.e.m.
Extended Data Figure 3 The therapy-induced secretome of sensitive cells overlaps significantly in melanoma and lung adenocarcinoma cells and appears after gene expression changes enriched for transcriptional regulators.
a, b, GO analysis (http://revigo.irb.hr) of gene expression changes after 6 h of vemurafenib treatment of A375 cells with spatial representation of enriched GO terms (a) and the molecular functions significantly affected (b). c, Heat map representing the expression levels of commonly up- and downregulated genes in vemurafenib-treated A375-derived xenograft tumours (5 days) and A375 cells in vitro (48 h). d, Principal component analysis of vemurafenib-sensitive Colo800 and UACC62 melanoma cells and crizotinib-sensitive H3122 lung adenocarcinoma cells treated in vitro with vehicle or vemurafenib or crizotinib for 48 h. e, Venn diagram indicating the overlap of genes in the extracellular region (GO:0005576) upregulated after 48 h of vemurafenib treatment in A375, Colo800 and UACC62 melanoma cell lines. f, Venn diagram indicating the overlap of genes in the extracellular region (GO:0005576) upregulated after 48 h of vemurafenib treatment in at least 2 out of 3 melanoma models and after 48 h of crizotinib treatment in the H3122 lung adenocarcinoma cell line. g, Venn diagram indicating the overlap of genes in the extracellular region (GO:0005576) downregulated after 48 h of vemurafenib treatment in A375, Colo800 and UACC62 melanoma cell lines. h, Venn diagram indicating the overlap of genes in the extracellular region (GO:0005576) downregulated after 48 h of vemurafenib treatment in at least 2 out of 3 melanoma models and after 48 h of crizotinib treatment in the H3122 lung adenocarcinoma cell line. P values calculated using a hypergeometric probability test.
Extended Data Figure 4 Vemurafenib treatment induces widespread changes in the intra-tumour immune cell composition and stromal cytokine composition in tumours regressing during targeted therapy.
a, b, FACS analysis of murine immune cell populations in A375-derived xenograft tumours treated with vehicle or vemurafenib for 5 days. a, b, Representative image (a) and quantification (b) of intra-tumour composition of indicated immune cell populations (vehicle, n = 4; vemurafenib, n = 6 tumours). c, d, Cytokine array of murine stroma-derived cytokines within A375-derived xenograft tumours treated with vehicle or vemurafenib for 5 days. Representative image (c) and quantification (d) of down- and upregulated cytokines (n = 4 tumours). P values calculated by a two-tailed Mann–Whitney U test. Data are mean and s.e.m.
Extended Data Figure 5 Targeted therapy induces downregulation of FRA1 in drug-sensitive tumour cells.
a, List of transcription factors predicted to regulate the vemurafenib-induced reactive secretome in A375 cells, and a heat map of the corresponding transcription factor gene expression levels in these cells. Red represents high, yellow medium and blue low relative expression on the colour scale. b, Immunoblotting of phosphorylated and total FRA1 protein levels in A375 and YUMM1.7 melanoma cell lines treated with vemurafenib for 24 h. c, Relative mRNA levels of FRA1 in H3122 cells treated with crizotinib (500 nM) and HCC827 treated with erlotinib (10 nM) at different time points (n = 4 technical replicates). d, Relative mRNA levels of FRA1 in A375R cells treated with vemurafenib at different time points (n = 4 technical replicates). e, Immunofluorescence staining of FRA1 (red) and DAPI (blue) in biopsies from melanoma patients before and after vemurafenib treatment (clinical information can be found in Extended Data Table 1). Original magnification, ×20.
Extended Data Figure 6 The secretome of melanoma cells with FRA1 knockdown stimulates proliferation and migration of A375R cells in vitro and in vivo.
a, Immunoblotting of phosphorylated and total FRA1 protein levels in A375 cells transduced with control shRNA, with or without additional vemurafenib treatment, or shRNAs targeting FRA1. b, Photon flux and representative BLI images of TGL-expressing A375R cells co-cultured with A375 cells expressing control shRNA (with or without vemurafenib treatment) or FRA1-targeting shRNAs after 7 days (n = 9 biological replicates). c, Relative number of A375R cells after 3 days in the presence of conditioned media derived from A375 cells transduced with control shRNA, with or without additional vemurafenib treatment, or FRA1 shRNAs (n = 3 biological replicates). d, Migration of A375R cells towards conditioned media derived from A375 cells transduced with control shRNA (with or without vemurafenib treatment) or FRA1 shRNAs using a Boyden chamber assay (shCtrl, n = 15; all other groups n = 10 FOV) e, Relative mRNA levels of selected secreted factors and transcription factors of A375 cells expressing control shRNA or an shRNA targeting FRA1 (shFRA1-1), treated with vehicle or vemurafenib (24 h). f, Bioluminescent signal of A375R-TGL cells 8 days after subcutaneous co-implantation with UACC62 cells expressing a control or an shRNA for FRA1 (shCtrl, n = 12; shFRA1, n = 20 tumours). Data are mean and s.e.m. P < 0.05, P < 0.01, P < 0.001, P < 0.0001, Student’s t-test.
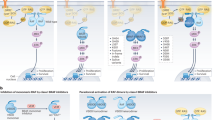
Extended Data Figure 7 The TIS includes upregulated positive regulators and a loss of negative regulators of the PI(3)K/AKT/mTOR pathway, which is activated in sensitive and resistant cells in vitro and in vivo.
a, b, Enriched biological processes (a) and inferred drug vulnerabilities (b) as determined by Ingenuity pathway analysis of gene expression data from vemurafenib-resistant A375R cells responding to signals from the reactive tumour microenvironment of a tumour regressing during targeted therapy in vivo (for experimental set-up see Fig. 1a and Methods). c, Left, immunoblotting of phosphorylated AKTS473 and phosphorylated ERK protein levels in A375 cells treated with vehicle or vemurafenib at different time points during the generation of conditioned media. Right, immunoblotting of phosphorylated AKTS473 and phosphorylated ERK protein levels in A375 cells after short-term exposure to conditioned media derived from A375 cells treated with vehicle or vemurafenib. d, Immunoblotting of phosphorylated AKTS473 and phosphorylated FRA1 protein levels in A375-derived xenograft tumours treated with vehicle or vemurafenib for 5 days. Normalized quantification of phospho-AKTS473/tubulin in the bottom panel. e, Immunoblotting of a range of pathway nodes in A375R cells treated with CM-vehicle or CM-vemurafenib, derived from A375 cells, for 15, 30, 60 or 120 min. f, Cancer cell-derived IGFBP3 levels (left) and murine stromal IGF1 levels (right) in A375-derived xenograft tumours treated with vehicle or vemurafenib for 5 days as determined by ELISA (n = 4 tumours) g, Cancer-cell-derived IGFBP3 levels in conditioned media from indicated melanoma cell lines treated with vehicle or vemurafenib as determined by ELISA (n = 3 technical replicates of conditioned media derived from at least two biological replicates). h, IGFBP3 levels in conditioned media derived from A375 cells expressing control shRNA or shRNAs targeting IGFBP3 (shIGFBP3-1 and -2) as determined by ELISA (n = 3 technical replicates). i, Immunoblotting of phosphorylated AKTS473 in A375R cells after incubation with conditioned media of A375 cells expressing control shRNA or shRNAs targeting IGFBP3. j, Phosphorylation status of AKTS473 in A375R cells after incubation for 15 min with conditioned media, IGF1 and IGFBP3 as indicated. k, Bioluminescent signal of A375R-TGL cells 10 days after co-implantation with A375 cells expressing a control shRNA or an shRNA targeting IGFBP3 (shIGFBP3-1) (n = 10 tumours). P values calculated by a two-tailed Mann–Whitney U test. Data are mean and s.e.m.
Extended Data Figure 8 Dual inhibition of RAF and the AKT/mTOR pathway blunts the effects of the regressing tumour environment on the resistant cell population.
a, Relative photon flux and representative BLI images of GFP/luciferase expressing A375R cells co-cultured with A375 cells and treated with vehicle, vemurafenib or the combination of vemurafenib and either MK2206 (AKTi, 2 μM) or BEZ235 (PI(3)K/mTORi, 300 nM) for 7 days (n = 2–3 biological replicates). b, Relative number of A375R cells after 3 days in the presence of CM-vehicle or CM-vemurafenib with additional BEZ235 (300 nM) (n = 3 biological replicates). c, Mice bearing tumours consisting of A375/A375R cells or A375R cells alone were treated with drugs as indicated. Bioluminescent signal of TGL-expressing A375R cells was determined on day 5 of treatment (n = 16, 16, 12, 12, 12 and 16 tumours, respectively). d, Mice bearing tumours consisting of unlabelled A375 cells were pre-treated for 3 days with drugs as indicated and 1 × 105 TGL-expressing A375R cells were inoculated in the arterial circulation. Drug treatment was continued and seeding of resistant cells to the primary tumour was quantified by BLI. Representative BLI images on the right (vehicle, n = 4; vemurafenib n = 10, vemurafenib plus BEZ235, n = 10 tumours). P values calculated by a two-tailed Mann–Whitney test. Data are mean and s.e.m.
Extended Data Figure 9 Characterization of cell lines in response to targeted therapy.
a–h, Relative survival of human melanoma cell lines (A375, Colo800 and UACC62) (a, c, e), and the murine melanoma cell line YUMM1.7 (g) and corresponding vemurafenib-resistant derivatives (A375R, Colo800R, UACC62R and YUMM1.7R) under increasing concentrations of vemurafenib. Immunoblotting of phosphorylated ERK protein levels in indicated melanoma cell lines in the presence of increasing concentrations of vemurafenib (b, d, f, h). i, Immunoblotting of phosphorylated ERK and phosphorylated AKTS473 protein levels in HCC827 lung adenocarcinoma cells in the presence of increasing concentrations of erlotinib. j, Immunoblotting of phosphorylated ERK protein levels in H3122 lung adenocarcinoma cells in the presence of increasing concentrations of crizotinib. k, Immunoblotting of protein levels of MET, EGFR, BRAF, PDGFRb, phosphorylated AKT and phosphorylated ERK in vemurafenib-sensitive and -resistant pairs of human melanoma cell lines (A375, Colo800 and UACC62).
Supplementary information
Supplementary Information
This file contains Supplementary Table 1. (XLSX 107 kb)
Rights and permissions
About this article
Cite this article
Obenauf, A., Zou, Y., Ji, A. et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature 520, 368–372 (2015). https://doi.org/10.1038/nature14336
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14336
This article is cited by
-
Apoptotic bodies: bioactive treasure left behind by the dying cells with robust diagnostic and therapeutic application potentials
Journal of Nanobiotechnology (2023)
-
Intratumor heterogeneity and cell secretome promote chemotherapy resistance and progression of colorectal cancer
Cell Death & Disease (2023)
-
SYK-mediated epithelial cell state is associated with response to c-Met inhibitors in c-Met-overexpressing lung cancer
Signal Transduction and Targeted Therapy (2023)
-
The mechanical phenotypic plasticity of melanoma cell: an emerging driver of therapy cross-resistance
Oncogenesis (2023)
-
Senescence-associated reprogramming induced by interleukin-1 impairs response to EGFR neutralization
Cellular & Molecular Biology Letters (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.