Abstract
Error-free repair of DNA double-strand breaks (DSBs) is achieved by homologous recombination (HR), and BRCA1 is an important factor for this repair pathway1. In the absence of BRCA1-mediated HR, the administration of PARP inhibitors induces synthetic lethality of tumour cells of patients with breast or ovarian cancers2,3. Despite the benefit of this tailored therapy, drug resistance can occur by HR restoration4. Genetic reversion of BRCA1-inactivating mutations can be the underlying mechanism of drug resistance, but this does not explain resistance in all cases5. In particular, little is known about BRCA1-independent restoration of HR. Here we show that loss of REV7 (also known as MAD2L2) in mouse and human cell lines re-establishes CTIP-dependent end resection of DSBs in BRCA1-deficient cells, leading to HR restoration and PARP inhibitor resistance, which is reversed by ATM kinase inhibition. REV7 is recruited to DSBs in a manner dependent on the H2AX–MDC1–RNF8–RNF168–53BP1 chromatin pathway, and seems to block HR and promote end joining in addition to its regulatory role in DNA damage tolerance6. Finally, we establish that REV7 blocks DSB resection to promote non-homologous end-joining during immunoglobulin class switch recombination. Our results reveal an unexpected crucial function of REV7 downstream of 53BP1 in coordinating pathological DSB repair pathway choices in BRCA1-deficient cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Roy, R., Chun, J. & Powell, S. N. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nature Rev. Cancer 12, 68–78 (2012)
Lee, J.-M., Ledermann, J. A. & Kohn, E. C. PARP Inhibitors for BRCA1/2 mutation-associated and BRCA-like malignancies. Ann. Oncol. 25, 32–40 (2013)
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005)
Lord, C. J. & Ashworth, A. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nature Med. 19, 1381–1388 (2013)
Ang, J. E. et al. Efficacy of chemotherapy in BRCA1/2 mutation carrier ovarian cancer in the setting of PARP inhibitor resistance: a multi-institutional study. Clin. Cancer Res. 19, 5485–5493 (2013)
Chun, A. C.-S., Kok, K.-H. & Jin, D.-Y. REV7 is required for anaphase-promoting complex-dependent ubiquitination and degradation of translesion DNA polymerase REV1. Cell Cycle 12, 365–378 (2013)
Jaspers, J. E. et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Discov 3, 68–81 (2013)
Listovsky, T. & Sale, J. E. Sequestration of CDH1 by MAD2L2 prevents premature APC/C activation prior to anaphase onset. J. Cell Biol. 203, 87–100 (2013)
Gan, G. N., Wittschieben, J. P., Wittschieben, B. Ø. & Wood, R. D. DNA polymerase zeta (pol zeta) in higher eukaryotes. Cell Res. 18, 174–183 (2008)
Hara, K. et al. Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase zeta and REV1. J. Biol. Chem. 285, 12299–12307 (2010)
Khalaj, M. et al. A missense mutation in Rev7 disrupts formation of Polζ, impairing mouse development and repair of genotoxic agent-induced DNA lesions. J. Biol. Chem. 289, 3811–3824 (2014)
Stap, J. et al. Induction of linear tracks of DNA double-strand breaks by alpha-particle irradiation of cells. Nature Methods 5, 261–266 (2008)
Fanning, E., Klimovich, V. & Nager, A. R. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 34, 4126–4137 (2006)
Sartori, A. A. et al. Human CtIP promotes DNA end resection. Nature 450, 509–514 (2007)
Bouwman, P. et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nature Struct. Mol. Biol. 17, 688–695 (2010)
Pierce, A. J., Johnson, R. D., Thompson, L. H. & Jasin, M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13, 2633–2638 (1999)
Bunting, S. F. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010)
Zimmermann, M. & de Lange, T. 53BP1: pro choice in DNA repair. Trends Cell Biol. 24, 108–117 (2014)
Chapman, J. R. et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell 49, 858–871 (2013)
Muramatsu, M. et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102, 553–563 (2000)
Schenten, D. et al. Pol ζ ablation in B cells impairs the germinal center reaction, class switch recombination, DNA break repair, and genome stability. J. Exp. Med. 206, 477–490 (2009)
Kikuchi, S., Hara, K., Shimizu, T., Sato, M. & Hashimoto, H. Structural basis of recruitment of DNA polymerase ζ by interaction between REV1 and REV7 proteins. J. Biol. Chem. 287, 33847–33852 (2012)
Evers, B. et al. Selective inhibition of BRCA2-deficient mammary tumor cell growth by AZD2281 and cisplatin. Clin. Cancer Res. 14, 3916–3925 (2008)
Fradet-Turcotte, A. et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499, 50–54 (2013)
Watanabe, S. et al. JMJD1C demethylates MDC1 to regulate the RNF8 and BRCA1-mediated chromatin response to DNA breaks. Nature Struct. Mol. Biol. 20, 1425–1433 (2013)
Doil, C. et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446 (2009)
Stewart, G. S. et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136, 420–434 (2009)
Stap, J. et al. Induction of linear tracks of DNA double-strand breaks by α-particle irradiation of cells. Nature Methods 5, 261–266 (2008)
de Jager, M. et al. DNA-binding and strand-annealing activities of human Mre11: implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Res. 29, 1317–1325 (2001)
Bouwman, P. et al. A high-throughput functional complementation assay for classification of BRCA1 missense variants. Cancer Discov. 3, 1142–1155 (2013)
Bartkova, J. et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 (2005)
Acknowledgements
We thank B. Gerritsen, P. Halonen, B. Morris, T. Halazonetis and O. Kallioniemi for advice on the DDR shRNA library, A. Gasparini and G. Borst for their assistance with the cone beam micro-irradiator, R. Kanaar for his RAD51 antibody, J. Jacobs for the pMSCV-GFP vector, and M. O’Connor for olaparib and AZD2461. This work was supported by the Netherlands Organization for Scientific Research (NWO-Toptalent to J.E.J. and NWO-VIDI to S.R.), the Dutch Cancer Society, CTMM Breast Care, the Swiss National Science Foundation, and the European Union (EU) FP6 Integrated Project CHEMORES and FP7 Project DDResponse. Work in J.R.C.’s group is funded by the Wellcome Trust. The work in the J.B.’s laboratories was funded by the Danish Cancer Society, the Danish Council for Independent Research, the Lundbeck Foundation and the Czech National Program of Sustainability. S.J.B. is funded by Cancer Research UK and an ERC Advanced Investigator Grant (RecMitMei) and is a Royal Society Wolfson Research Merit Award Holder.
Author information
Authors and Affiliations
Contributions
G.X. and S.R. designed the study, performed experiments and wrote the manuscript; I.B. and D.C.v.G. designed and performed the RPA foci analysis; J.R.C., A.S. and S.J.B. performed and planned the CSR assay and RIF1-associated data; J.C. and J.Y. designed and performed the experiments using MEFs; J.E.J., A.K. and W.S. assisted with the mouse intervention studies; E.G. established the in vivo RAD51 analysis that K.J. and B.v.d.B. quantified; P.H.N.C. designed and M.B. helped in generating the REV7 mutants; Pe.B., M.P. and J.J. helped in designing the shRNA screen and performed experiments using mES cells; M.M. performed the laser stripe assays, and K.W. performed co-immunoprecipitations. D.W. helped to visualize GFP–REV7 recruitment; M.N., I.d.R. and J.d.R. carried out the RNA sequencing (RNA-seq) data analysis, Jirina B. established, carried out and evaluated the REV7 immunohistochemistry, P.C.S. helped with the analysis of the immunohistochemistry data, Jiri B. and Pi.B. advised on experiments and manuscript revisions, and all authors discussed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
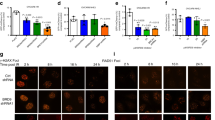
Extended Data Figure 1 Loss of Rev7 causes PARPi resistance in vitro and in vivo.
a, Quantification of Rev7 transcript levels in KB1P-B11 cells transduced with Rev7-targeting shRNAs or the vector control. Hprt was used as a control for transcript expression. The data represent mean ± s.d. b, c, Cell proliferation rates in KB1P-G3 (b) or KB1P-B11 (c) cells analysed using the MTT assay. d–g, Long-term clonogenic survival assays and quantification of KB1P-G3 (d, f) or KB1P-B11 (e, g) cells transduced with the indicated constructs and treatments. All the groups were normalized by the absorbance of the vector control. The data represent mean ± s.d. h, Quantification of the real colony numbers from the short-term clonogenic survival assay of KB1P-G3 cells with or without Rev7 loss exposed to olaparib. i, REV7 protein levels were determined by western blotting of lysates derived from KB1P-G3 cells transduced with the indicated constructs. j, Overall survival of mice with KB1P-B11-derived Rev7-depleted or control tumours treated with one regimen of 50 mg kg−1 olaparib daily for 28 days or left untreated. The P value was calculated using the log-rank test. k, l, Relative tumour growth of individual KB1P-G3- (k) and KB1P-B11- (l) derived Rev7-depleted or control tumours treated with one regimen of 50 mg kg−1 olaparib daily for 28 days or left untreated.
Extended Data Figure 2 Loss of REV7 causes olaparib resistance in BRCA1-deficient SUM149PT cells.
a, Western blotting analysis of REV7 or 53BP1 expression in SUM149PT cells transduced with REV7- or 53BP1-targeting hairpins or the vector control. b, Example of a long-term clonogenic survival assay using the indicated hairpins and olaparib concentrations. c, Quantification of the clonogenic assays using absorbance of crystal violet at 590 nm. The data represent mean ± s.d. All the groups were normalized by the absorbance of the vector control and showed significant differences to the control (P < 0.01, t-test).
Extended Data Figure 3 Rev1 or Rev3 inhibition and PARPi sensitivity of Brca1−/− p53−/− mammary tumour cells.
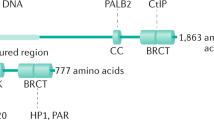
a, b, Quantification of Rev1 (a) or Rev3 (b) transcript levels in KB1P-G3 cells transduced with Rev1- or Rev3-targeting shRNAs or the vector control. Hprt was used as a control for transcript expression. The data represent the mean ± s.d. c, Long-term clonogenic survival assays of KB1P-G3 cells exposed to the indicated PARP inhibitors. d, Quantification of the clonogenic assay by determining the absorbance of crystal violet at 590 nm. All the groups were normalized by the absorbance of the vector control. The data represent the mean ± s.d. e, f, Quantification of Rev7 transcript (e) or protein (f) levels in KB1P-G3 cells transduced with Rev7-targeting shRNAs or the vector control. Hprt was used as a control for transcript expression; α-tubulin as a control for protein expression. The data represent mean ± s.d. g, GFP-tagged REV7 mutants recruitment to sites of DNA damage was observed 5 min after 405 nm laser exposure (0.99 mW, 60% laser power, 50 s) in KB1P-B11 cells. Scale bar, 10 μm.
Extended Data Figure 4 REV7 recruitment to the DNA damage sites in human cells.
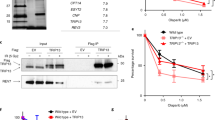
a–c, Human REV7 recruitment to sites of laser-induced DNA damage was analysed in U2OS cells transfected with siRNAs targeting RNF8 (siRNF8), RNF168 (siRNF168) (a), 53BP1 (si53BP1) (c) and GFP (siGFP). RNF8 and RNF168 protein levels were determined by western blotting (b) using lysates derived from U2OS cells transfecting with the indicated siRNAs. d, For the quantification of the REV7 signal within laser-induced DNA damage stripes, a minimum of 100 striped (that is, γH2AX-positive) cells were analysed for the presence of the REV7 signal in two independent experiments. Scale bars, 50 μm. e, RAD51 focus (red) formation in KB1P-B11 cells before and 5 h after 10 Gy ionizing radiation. Scale bar, 10 μm. f, Quantification of RAD51 foci in KB1P-B11 cells in the presence or absence of REV7 depletion. At least 150 cells were analysed per group in three independent experiments each. Error bars indicate s.d.; IR denotes 5 h after 10 Gy ionizing radiation. g, Western blotting analysis of REV7-depleted KB1P-G3 cells transfected with human REV7–GFP or Rev7-shRNA-resistant mouse REV7–GFP fusion proteins. h, Same as in e and f using the ATM inhibitor KU55933 with or without IR (5 h after 10 Gy ionizing radiation). i, Long-term clonogenic survival assay of KB1P-G3 cells exposed to olaparib in the presence or absence of KU55933 pre-treatment.
Extended Data Figure 5 Loss of Rev7 does not cause PARPi resistance in Brca2−/− p53−/− or p53−/− mammary tumour cells in vitro.
a, b, Quantification of Rev7 transcript levels in Brca2−/− p53−/− (KB2P-1.21 or KB2P-3.4) cells transduced with Rev7-targeting shRNAs or the vector control. Hprt was used as a control for transcript expression. The data represent the mean ± s.d. c–f, Long-term clonogenic survival assays and quantification of KB2P-1.21 or KB2P-3.4 cells with or without Rev7 depletion exposed to the indicated treatments. All the groups were normalized by the absorbance of the vector control. The data represent mean ± s.d. g, Quantification of Rev7 transcript levels in p53−/− (KP3.33) cells transduced with the indicated constructs. Hprt was used as a control for transcript expression and the data represent the mean ± s.d. h, i, Long-term clonogenic survival assays and quantification of KP3.33 cells exposed to the indicated treatments. The data represent the mean ± s.d.
Extended Data Figure 6 Rev7 loss promotes end resection at DSBs in BRCA1-deficient cells after ionizing radiation.
a, Quantification of RPA-positive α tracks in KB1P-B11 cells 1 or 2 h after irradiation with α particles. b, Quantification of RPA- and 53BP1-positive α tracks in KB1P-B11 cells transfected with non-targeting control siRNAs or siRNAs against Ctip. c, Cell cycle analysis (BrdU incorporation and propidium iodine labelling) of KB1P-B11 cells transduced with the indicated constructs and siRNAs. d, e, Quantification of Rev7 transcript (d) or protein (e) levels in BRCA1-deficient mES cells transduced with Rev7-targeting shRNAs or the vector control. Hprt was used as a control for transcript expression, α-tubulin as a control for protein expression. The data represent mean ± s.d. f, Representative images of surviving colonies of Brca1−/− mES cells transduced with an empty vector control or Rev7-targeting shRNAs. g, Quantification of colony formation normalized to the vector control. h, Quantification of RAD51 foci in Brca1−/− mES cells that were transduced with the indicated constructs.
Extended Data Figure 7 REV7 loss frequently occurs in triple-negative breast cancer.
a, b, Quantification of human REV7 transcript levels (a) and protein levels (b) in U2OS cells transduced with indicated constructs. Two different pairs of primers for REV7 or HPRT were used for the quantification of REV7 transcript levels. c–e, Examples of aberrantly reduced REV7 protein expression in triple-negative human breast carcinomas. Immunohistochemical detection of REV7 in human breast carcinomas shows moderate to high nuclear expression in normal human breast tissue (data not shown), and most invasive breast tumours (c). Aberrant reduction of REV7 with less than 70% of cancer cells that show nuclear positivity (d, e) was observed in 18 out of 50 cases.
Extended Data Figure 8 REV7 is a downstream effector of 53BP1.
a, Quantification of 53BP1 foci in KB1P-G3 cells in the presence or absence of REV7 depletion. At least 100 cells were analysed per group in three independent experiments each. Error bars indicate s.d.; IR denotes 5 h after 10 Gy ionizing radiation. b, REV7 or 53BP1 protein levels were determined by western blotting of lysates derived from MEF cells transduced with the indicated control (CNTL) or Rev7- and 53bp1-targeting shRNA constructs. c, d, RIF1 foci formation (c) after neocarzinostatin (NCS) treatment and quantification of RIF1 foci (d) in MEF cells in the presence or absence of REV7 or 53BP1 depletion. e, Flag pull-downs were performed from 2 mg lysate prepared from 53bp1−/−, 53bp1−/− plus 53BP1 and 53bp1−/− plus 53BP120AQ MEFs after mock or neocarzinostatin treatment. Control, 53BP1 and 53BP120AQ immunoprecipitates were incubated in HeLa nuclear extract (HNE, 2 mg) and then eluted with triple-Flag peptide. HA, haemagglutinin.
Extended Data Figure 9 The effect of REV7 inhibition on CSR after antigenic stimulation of CH12 cells.
a, Rev7 messenger RNA levels determined by qRT–PCR were normalized against β-actin (Actb) transcripts in the indicated shRNA-transduced CH12 cell lines. The data represent the mean ± s.e.m. from two primer sets specific for Rev7 transcript. CNTL, control. b, 53BP1 protein of each group normalized to vector-transduced cells (CH12) was analysed by western blotting. c, IgH μ and α germ-line transcripts (GLT) and Aid mRNA were estimated by semi-quantitative RT–PCR using twofold serial dilutions of cDNA made from indicated CH12 cell lines 40 h after stimulation. Hprt was used as a control for transcript expression. d, Representative flow cytometric profiles of shRNA-transduced CH12 B cells stained with anti-IgA antibody 40 h after stimulation with the indicated cytokines. e, Cells (CH12) were labelled with CFSE immediately before cytokine stimulation as in Fig. 4d, and cell proliferation was assessed by flow cytometry at indicated time points. f, Quantification of CSR to IgA of shRNA-transduced CH12 cells 40 h after stimulation with CD40 antibody, IL-4 and TGF-β1 (CIT). Data represent the mean ± s.d. from two independent experiments performed in triplicate.
Supplementary information
Supplementary Information
This file contains Supplementary Table 1. (XLS 992 kb)
Rights and permissions
About this article
Cite this article
Xu, G., Chapman, J., Brandsma, I. et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 521, 541–544 (2015). https://doi.org/10.1038/nature14328
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14328
This article is cited by
-
UFL1 triggers replication fork degradation by MRE11 in BRCA1/2-deficient cells
Nature Chemical Biology (2024)
-
Leveraging the replication stress response to optimize cancer therapy
Nature Reviews Cancer (2023)
-
Transmembrane nuclease NUMEN/ENDOD1 regulates DNA repair pathway choice at the nuclear periphery
Nature Cell Biology (2023)
-
Synergistic effect of combined PI3 kinase inhibitor and PARP inhibitor treatment on BCR/ABL1-positive acute lymphoblastic leukemia cells
International Journal of Hematology (2023)
-
Distinct roles of treatment schemes and BRCA2 on the restoration of homologous recombination DNA repair and PARP inhibitor resistance in ovarian cancer
Oncogene (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.