Abstract
Tissue homeostasis is achieved through a balance of cell production (growth) and elimination (regression)1,2. In contrast to tissue growth, the cells and molecular signals required for tissue regression remain unknown. To investigate physiological tissue regression, we use the mouse hair follicle, which cycles stereotypically between phases of growth and regression while maintaining a pool of stem cells to perpetuate tissue regeneration3. Here we show by intravital microscopy in live mice4,5,6 that the regression phase eliminates the majority of the epithelial cells by two distinct mechanisms: terminal differentiation of suprabasal cells and a spatial gradient of apoptosis of basal cells. Furthermore, we demonstrate that basal epithelial cells collectively act as phagocytes to clear dying epithelial neighbours. Through cellular and genetic ablation we show that epithelial cell death is extrinsically induced through transforming growth factor (TGF)-β activation and mesenchymal crosstalk. Strikingly, our data show that regression acts to reduce the stem cell pool, as inhibition of regression results in excess basal epithelial cells with regenerative abilities. This study identifies the cellular behaviours and molecular mechanisms of regression that counterbalance growth to maintain tissue homeostasis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bergmann, A. & Steller, H. Apoptosis, stem cells, and tissue regeneration. Sci. Signal. 3, re8 (2010)
Poon, I. K., Lucas, C. D., Rossi, A. G. & Ravichandran, K. S. Apoptotic cell clearance: basic biology and therapeutic potential. Nature Rev. Immunol. (2014)
Müller-Röver, S. et al. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J. Invest. Dermatol. 117, 3–15 (2001)
Rompolas, P. et al. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature 487, 496–499 (2012)
Rompolas, P., Mesa, K. R. & Greco, V. Spatial organization within a niche as a determinant of stem-cell fate. Nature (2013)
Deschene, E. R. et al. β-Catenin Activation Regulates Tissue Growth Non–Cell Autonomously in the Hair Stem Cell Niche. Science 343, 1353–1356 (2014)
Lindner, G. et al. Analysis of apoptosis during hair follicle regression (catagen). Am. J. Pathol. 151, 1601 (1997)
Rogers, G. E. Hair follicle differentiation and regulation. Int. J. Dev. Biol. 48, 163–170 (2004)
Li, J. L. et al. Intravital multiphoton imaging of immune responses in the mouse ear skin. Nature Protocols 7, 221–234 (2012)
Monks, J. et al. Epithelial cells as phagocytes: apoptotic epithelial cells are engulfed by mammary alveolar epithelial cells and repress inflammatory mediator release. Cell Death Differ. 12, 107–114 (2005)
Monks, J., Smith-Steinhart, C., Kruk, E. R., Fadok, V. A. & Henson, P. M. Epithelial cells remove apoptotic epithelial cells during post-lactation involution of the mouse mammary gland. Biol. Reprod. 78, 586–594 (2008)
Hsu, Y.-C., Pasolli, H. A. & Fuchs, E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell 144, 92–105 (2011)
Choi, Y. S. et al. Distinct functions for Wnt/β-catenin in hair follicle stem cell proliferation and survival and interfollicular epidermal homeostasis. Cell Stem Cell 13, 720–733 (2013)
Foitzik, K. et al. Control of murine hair follicle regression (catagen) by TGF-β1 in vivo. FASEB J. 14, 752–760 (2000)
Larsson, J. et al. TGF-β signalling-deficient hematopoietic stem cells have normal self-renewal and regenerative ability in vivo despite increased proliferative capacity in vitro. Blood 102, 3129–3135 (2003)
Rahmani, W. et al. Hair follicle mesenchymal stem cells regenerate the dermal sheath, replenish the dermal papilla and specify hair type. Dev. Cell 31, 543–558 (2014)
Tumbar, T. et al. Defining the epithelial stem cell niche in skin. Science 303, 359–363 (2004)
Rendl, M., Lewis, L. & Fuchs, E. Molecular dissection of mesenchymal–epithelial interactions in the hair follicle. PLoS Biol. 3, e331 (2005)
Vaezi, A., Bauer, C., Vasioukhin, V. & Fuchs, E. Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev. Cell 3, 367–381 (2002)
Harada, N. et al. Intestinal polyposis in mice with a dominant stable mutation of the β‐catenin gene. EMBO J. 18, 5931–5942 (1999)
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007)
Harfe, B. D. et al. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517–528 (2004)
Clausen, B. E., Burkhardt, C., Reith, W., Renkawitz, R. & Förster, I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8, 265–277 (1999)
Jung, S. et al. Analysis of fractalkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20, 4106–4114 (2000)
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature Neurosci. 13, 133–140 (2010)
Zito, G. et al. Spontaneous tumour regression in keratoacanthomas is driven by Wnt/retinoic acid signalling cross-talk. Nature Commun. 5, 3543 (2014)
Greco, V. et al. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell 4, 155–169 (2009)
Clavel, C. et al. Sox2 in the dermal papilla niche controls hair growth by fine-tuning BMP signaling in differentiating hair shaft progenitors. Dev. Cell 23, 981–994 (2012)
Acknowledgements
We thank E. Fuchs for the K14-H2BGFP, Lef1-RFP and K14-GFPActin mice; M. Taketo for the Cttnb1fl(Ex3)/+ mice; A. Horwich and A. Mesa for critical feedback; M. Rendl for technical advice; M. Graham and X. Liu for technical support with electron microscopy; D. Egli for the H2BmCherry construct; and T. Nottoli for generating the K14-H2BmCherry mouse line. K.R.M. and S.B. were supported by the National Institutes of Health (NIH) Predoctoral Program in Cellular and Molecular Biology, grant no. 5T32 GM007223. K.R.M. is currently a National Science Foundation (NSF) Graduate Research Fellow. This work is supported by the American Cancer Society, grant no. RSG-12-059-02; Yale Spore Grant National Cancer Institute, NIH, grant no. 2P50CA121974; the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), NIH, grant no. 1R01AR063663-01; and by The New York Stem Cell Foundation. V.G. is a New York Stem Cell Foundation–Robertson Investigator. P.R. is a New York Stem Cell Foundation–Druckenmiller Fellow. A.M.H. is supported by NIAMS Rheumatic Diseases Research Core Centers grant no. 5 P30 AR053495-07. K.B.B. was supported by the NSF. The NSF had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The views presented here are not those of the NSF and represent solely the views of the authors.
Author information
Authors and Affiliations
Contributions
K.R.M. and V.G. designed experiments and wrote the manuscript; K.R.M. performed the experiments and analysed the data. P.R. generated the K14-H2BmCherry mouse line and assisted with two-photon time-lapse imaging. G.Z. and P.M. performed immunofluorescence. S.B. performed skin whole-mount staining. T.Y.S. assisted with technical aspects. K.R.M., D.G.G. and A.M.H. performed three-dimensional imaging analysis. K.B.B. helped with data analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Hair follicle regeneration cycle
Hair follicle growth (Anagen) is characterized by downward expansion and generation of several epithelial layers. The most external layer, the outer root sheath (ORS), consists of relatively undifferentiated basal epithelial cells. Inner layers are generated by a committed progenitor pool, the matrix, which gives rise to several differentiated layers including: companion layer, inner root sheath (IRS) and hair shaft. After growth, the majority of the newly formed layers are lost during the regression phase (Catagen), leaving a small surviving fraction of cells that reconstitute a new stem cell/progeny (Bulge/Hair Germ) compartment at the rest phase (Telogen).
Extended Data Figure 2 Hair follicle inner layers resist apoptosis and continue upward terminal differentiation.
a, Upward movement of hair follicle inner layers during growth and regression. Single optical sections show upward collective movement of inner layers relative to surrounding basal cells at two time points 130 min apart. Compare the position of labelled cells and dashed line of basal (red) to inner layers (yellow). b, Upward movement of hair shaft during regression. Optical sections of top view (epidermis) and side view (hair follicle) at two time points, 1 day apart. Note the extrusion of hair shafts from regressing hair follicles. Observations shown represent n = 3 mice. c, Companion layer lineage tracing during regression. Representative example of matrix progenitors of the companion layer traced during regression in Lgr5-CreER;tdTomato;K14-H2BGFP mice (n = 20 or 7 lineages, in 4 mice). d, Terminal differentiation of inner layer progenitor cells. Representative example of single-cell lineages (n = 35 or 9 lineages, in 3 mice) traced during the initial transition of hair follicle growth to regression in Shh-CreER;tdTomato;K14-H2BGFP mice. Scale bars, 25 µm.
Extended Data Figure 3 Apoptotic bodies are cleared by neighbouring basal epithelial cells.
a, Toluidine-blue-stained section of regressing hair follicles used for ultrastructure analysis. b, Electron micrograph illustrating multiple apoptotic bodies (red arrowheads) present in hair follicle basal epithelium, but absent in inner layers. c, Electron micrograph showing a hair follicle in regression (white dashed line). d, Electron micrograph showing the restriction of apoptotic bodies (red arrowheads) and phagocytic activity to the basal epithelium. e, High-magnification electron micrograph with immune-gold labelling for GFP protein expressed by K14-H2BGFP+ cells. Positive GFP labelling is present in both apoptotic bodies (Ap) and phagocytic basal epithelial nuclei (n). Observations shown represent n = 2 mice. Scale bars, 25 µm unless otherwise indicated.
Extended Data Figure 4 Professional phagocytes are not present in regressing hair follicles.
a, Professional phagocytes do not enter regressing hair follicles. Single optical sections showing absence of myeloid populations inside the hair follicle 2.5 h after epithelial cell death (arrowhead) in LysM-Cre;tdTomato;K14-H2BGFP mice. b, Immunofluorescent staining of myeloid populations in skin during hair follicle regression. DAPI, blue; CD11b, red; P-cadherin, green. Observations shown represent n = 4 mice. Scale bar, 25 µm.
Extended Data Figure 5 Wnt/β-catenin activity is restricted to the inner layers during regression.
a, b, Immunohistochemistry (a) and immunofluorescent (b) staining highlighting active (nuclear) β-catenin of hair follicle inner layers (dashed line) at the onset of regression. c, Immunofluorescent staining of the Wnt/β-catenin target gene, Lef1, during hair follicle regression. DAPI, blue; Lef1, red; P-cadherin, green. Asterisk indicates mesenchymal dermal papilla. Observations shown represent n = 2 mice. Scale bars, 50 µm.
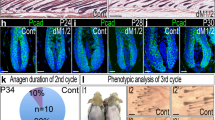
Extended Data Figure 6 Late and partial mesenchymal dermal papilla removal does not affect hair follicle regression.
a, Sequential revisits of hair follicles after dermal papilla (DP) ablation during late regression. b, Box plot quantification of hair follicle length immediately after ablation, 4 days and 11 days after dermal papilla ablation (n = 20 follicles, in 4 mice; error bars represent maximum and minimum). c, Sequential revisits of hair follicles after partial dermal papilla ablation during early regression (n = 12 follicles, in 3 mice). d, Schematic illustration of the results from mesenchymal dermal papilla ablation experiments. Dermal papilla ablation during early regression results in failed elimination of the basal epithelium, while the inner layers continue upward in terminal differentiation, yet dermal papilla ablation during late regression does not impair hair follicle regression. Asterisk indicates auto-fluorescence from the two-photon laser. NS, not significant; P < 0.05, mean ± s.d. Scale bars, 25 µm.
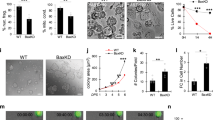
Extended Data Figure 7 Characterization of TGF-β pathway in mesenchymal dermal papilla and basal epithelial cell populations during regression.
a, Schematic of skin digestion and cell isolation with representative images before and after tissue digestion in K14-H2BGFP;Lef1-RFP mice. b, Representative fluorescent-activated cell sorting (FACS) scheme for isolating mesenchymal dermal papilla (DP; RFP+, CD34−, CD45−, CD117−, integrin-α9+) and enriched hair follicle basal epithelium (RFP−, GFPHigh) cells. c, Validation of mesenchymal dermal-papilla-sorted population enrichment by Sox2 expression. d, Validation of basal-epithelial-sorted population enrichment by keratin 14 (K14) expression. e, TGF-β ligand 2 and 3 expression in the mesenchymal dermal papilla throughout the hair cycle. TGF-β1 expression in basal epithelium, mesenchymal dermal papilla, and all sorted cells during regression. f, Differential expression of TGF-β target genes: Smad7, Tmeff1, p15INK4B (also known as Cdkn2b) and in the hair follicle basal epithelium throughout the hair cycle (mean ± s.d.; n = 3 technical replicates). Scale bars, 100 µm. *P < 0.05, **P < 0.01, ***P < 0.001.
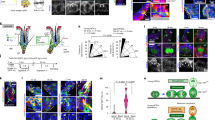
Extended Data Figure 8 Local TGF-β activation during regression and validation of Cre-induced loss of TGF-βR1 expression.
a, TGF-β activation shown by immunofluorescent staining of pSmad2 during the transition from hair follicle growth to regression. DAPI, blue; pSmad2, red; P-cadherin, green. Observations shown represent n = 4 mice. b, Representative FACS scheme for isolating tdTomato-Cre-reporter-positive basal epithelial cells (tdTomato+, GFPHigh) from Tgfbr1fl/fl and Tgfbr1fl/+ mice. c, Expression of TGF-βR1 and the TGF-β target gene, Smad7, in Cre-recombined basal epithelial cells from Tgfbr1fl/fl and Tgfbr1fl/+ mice (P < 0.01, mean ± s.d.; n = 3 technical replicates). Scale bar, 50 µm.
Extended Data Figure 9 Extrinsic induction of hair follicle regression dictates the regenerative (stem cell) pool.
Crosstalk with the mesenchymal niche during regression results in localized TGF-β activation, promoting a spatially restricted gradient of cell death in the basal epithelium. Clearance of apoptotic cells by neighbouring basal epithelial cells results in a limited pool of surviving stem cells. Inhibition of this regression process results in excessive amounts of basal epithelial cells capable of fuelling a new round of growth when in contact with the mesenchymal dermal papilla.
Supplementary information
Supplementary Table 1
This table shows a list of primers used for RT-qPCR. (PDF 107 kb)
Hair follicle regression captured in vivo
Time-lapse recording of regressing hair follicles as seen using a K14-H2BGFP reporter in a live mouse by two-photon laser scanning microscopy (hh:mm). (MOV 2405 kb)
Upward movement of hair follicle inner layers during regression
Time-lapse recording of inner layers as seen using a K14-H2BGFP reporter. Note the relative upward displacement of the inner layers (centrally located) (hh:mm). Also see Fig 1b. (MOV 1162 kb)
Basal epithelial cell death captured in vivo
Time-lapse recording of basal epithelial cell death as seen by nuclear fragmentation using a K14-H2BGFP reporter (hh:mm). Note the relocation of nuclear fragments around neighboring epithelial nuclei. Also see Fig 1d. (MOV 289 kb)
Serial optical sections capture cellular dynamics during regression
Gallery view of a time-lapse recording highlights the complexity of the regression process and illustrates how several cellular behaviors and events occur in parallel using K14-H2BmCherry and K14-GFPActin reporters. Note that this view captures both an apoptotic event (in Row 3) as well as a phagocytic event (in Row 2). (MOV 7531 kb)
Live imaging of the sequential cellular behaviors of epithelial self-clearance
Time-lapse recording of multiple apoptotic bodies from a single apoptotic basal cell being engulfed by neighboring basal epithelial cells using K14-GFPActin and K14-H2BmCherry reporters. Note that the apoptotic body originated from an apoptotic cell located 8µm below the site of engulfment. The apoptotic body is internalized by a neighboring epithelial cell as shown in both the (x,y) and (x,z) views. Also see Fig. 1g. (MOV 1511 kb)
3D tracking of epithelial phagocytosis of an apoptotic body captured in vivo
3D tracking of three apoptotic bodies formed from a single apoptotic basal epithelial cell as visualized by K14-GFPActin and K14-H2BmCherry reporters. Note the individual movement and divergent paths of the apoptotic bodies from the apoptotic cell (red) toward one of the phagocytic cells (green). (MOV 3962 kb)
LysMCre labeled cells do not enter regressing hair follicles
Time-lapse recording of a hair follicle in regression with surrounding LysMCre+ dermal populations (red) as seen using LysM-Cre;tdTomato reporter, in addition to the K14-H2BGFP reporter. Note that the nuclear fragments (green) are retained in the hair follicle. Also see Fig. 2a and Extended Data Fig. 4a. (MOV 1538 kb)
CX3CR1-GFP+ cells do not engulf apoptotic epithelial cells from regressing hair follicles
Time-lapse recording of a hair follicle in regression with surrounding myeloid populations (green) as seen using CX3CR1-GFP, in addition to the K14-H2BmCherry reporter. Note that the epithelial nuclear fragments (red) are retained in the regressing hair follicle. (MOV 4109 kb)
Basal epithelial cell death at the mesenchymal DP interface during early regression
Time-lapse recording of a hair follicle in early regression as seen using the K14-H2BGFP and Lef1-RFP reporters. Note the contact between dying basal epithelial cells (green nuclei) and DP (red cells) during regression (hh:mm). (MOV 1777 kb)
Rights and permissions
About this article
Cite this article
Mesa, K., Rompolas, P., Zito, G. et al. Niche-induced cell death and epithelial phagocytosis regulate hair follicle stem cell pool. Nature 522, 94–97 (2015). https://doi.org/10.1038/nature14306
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14306
This article is cited by
-
Local and systemic mechanisms that control the hair follicle stem cell niche
Nature Reviews Molecular Cell Biology (2024)
-
An optimized force-triggered density gradient sedimentation method for isolation of pelage follicle dermal papilla cells from neonatal mouse skin
Stem Cell Research & Therapy (2023)
-
Lrig1-expressing epidermal progenitors require SCD1 to maintain the dermal papilla niche
Scientific Reports (2023)
-
Hdac1 and Hdac2 regulate the quiescent state and survival of hair-follicle mesenchymal niche
Nature Communications (2023)
-
Extracellular Vesicles: Therapeutic Potential in Central Nervous System Trauma by Regulating Cell Death
Molecular Neurobiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.