Abstract
Influenza virus polymerase uses a capped primer, derived by ‘cap-snatching’ from host pre-messenger RNA, to transcribe its RNA genome into mRNA and a stuttering mechanism to generate the poly(A) tail. By contrast, genome replication is unprimed and generates exact full-length copies of the template. Here we use crystal structures of bat influenza A and human influenza B polymerases (FluA and FluB), bound to the viral RNA promoter, to give mechanistic insight into these distinct processes. In the FluA structure, a loop analogous to the priming loop of flavivirus polymerases suggests that influenza could initiate unprimed template replication by a similar mechanism. Comparing the FluA and FluB structures suggests that cap-snatching involves in situ rotation of the PB2 cap-binding domain to direct the capped primer first towards the endonuclease and then into the polymerase active site. The polymerase probably undergoes considerable conformational changes to convert the observed pre-initiation state into the active initiation and elongation states.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Resa-Infante, P., Jorba, N., Coloma, R. & Ortin, J. The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol. 8, 207–215 (2011)
Fodor, E. The RNA polymerase of influenza a virus: mechanisms of viral transcription and replication. Acta Virol. 57, 113–122 (2013)
Ulmanen, I., Broni, B. A. & Krug, R. M. Role of two of the influenza virus core P proteins in recognizing cap 1 structures (m7GpppNm) on RNAs and in initiating viral RNA transcription. Proc. Natl Acad. Sci. USA 78, 7355–7359 (1981)
Blass, D., Patzelt, E. & Kuechler, E. Identification of the cap binding protein of influenza virus. Nucleic Acids Res. 10, 4803–4812 (1982)
Guilligay, D. et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nature Struct. Mol. Biol. 15, 500–506 (2008)
Dias, A. et al. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458, 914–918 (2009)
Yuan, P. et al. Crystal structure of an avian influenza polymerase PAN reveals an endonuclease active site. Nature 458, 909–913 (2009)
Pritlove, D. C., Poon, L. L., Fodor, E., Sharps, J. & Brownlee, G. G. Polyadenylation of influenza virus mRNA transcribed in vitro from model virion RNA templates: requirement for 5′ conserved sequences. J. Virol. 72, 1280–1286 (1998)
Nemeroff, M. E., Barabino, S. M., Li, Y., Keller, W. & Krug, R. M. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1, 991–1000 (1998)
Bier, K., York, A. & Fodor, E. Cellular cap-binding proteins associate with influenza virus mRNAs. J. Gen. Virol. 92, 1627–1634 (2011)
Jorba, N., Coloma, R. & Ortin, J. Genetic trans-complementation establishes a new model for influenza virus RNA transcription and replication. PLoS Pathog. 5, e1000462 (2009)
York, A., Hengrung, N., Vreede, F. T., Huiskonen, J. T. & Fodor, E. Isolation and characterization of the positive-sense replicative intermediate of a negative-strand RNA virus. Proc. Natl Acad. Sci. USA 110, E4238–E4245 (2013)
Nie, Y., Bellon-Echeverria, I., Trowitzsch, S., Bieniossek, C. & Berger, I. Multiprotein complex production in insect cells by using polyproteins. Methods Mol. Biol. 1091, 131–141 (2014)
Lee, Y. S. & Seong, B. L. Nucleotides in the panhandle structure of the influenza B virus virion RNA are involved in the specificity between influenza A and B viruses. J. Gen. Virol. 79, 673–681 (1998)
Pflug, A., Guilligay, D., Reich, S. & Cusack, S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature http://dx.doi.org/10.1038/nature14008 (this issue)
Flick, R., Neumann, G., Hoffmann, E., Neumeier, E. & Hobom, G. Promoter elements in the influenza vRNA terminal structure. RNA 2, 1046–1057 (1996)
Pritlove, D. C., Poon, L. L., Devenish, L. J., Leahy, M. B. & Brownlee, G. G. A hairpin loop at the 5′ end of influenza A virus virion RNA is required for synthesis of poly(A)+ mRNA in vitro. J. Virol. 73, 2109–2114 (1999)
Newcomb, L. L. et al. Interaction of the influenza a virus nucleocapsid protein with the viral RNA polymerase potentiates unprimed viral RNA replication. J. Virol. 83, 29–36 (2009)
Moeller, A., Kirchdoerfer, R. N., Potter, C. S., Carragher, B. & Wilson, I. A. Organization of the influenza virus replication machinery. Science 338, 1631–1634 (2012)
Butcher, S. J., Grimes, J. M., Makeyev, E. V., Bamford, D. H. & Stuart, D. I. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410, 235–240 (2001)
Lescar, J. & Canard, B. RNA-dependent RNA polymerases from flaviviruses and Picornaviridae. Curr. Opin. Struct. Biol. 19, 759–767 (2009)
Gong, P. & Peersen, O. B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl Acad. Sci. USA 107, 22505–22510 (2010)
Deng, T., Vreede, F. T. & Brownlee, G. G. Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. J. Virol. 80, 2337–2348 (2006)
Mosley, R. T. et al. Structure of hepatitis C virus polymerase in complex with primer-template RNA. J. Virol. 86, 6503–6511 (2012)
Plotch, S. J., Bouloy, M., Ulmanen, I. & Krug, R. M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23, 847–858 (1981)
Reguera, J., Weber, F. & Cusack, S. Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog. 6, e1001101 (2010)
Sikora, D., Rocheleau, L., Brown, E. G. & Pelchat, M. Deep sequencing reveals the eight facets of the influenza A/HongKong/1/1968 (H3N2) virus cap-snatching process. Sci. Rep. 4, 6181 (2014)
Datta, K., Wolkerstorfer, A., Szolar, O. H., Cusack, S. & Klumpp, K. Characterization of PA-N terminal domain of Influenza A polymerase reveals sequence specific RNA cleavage. Nucleic Acids Res. 441, 349–353 (2013)
Hagen, M., Tiley, L., Chung, T. D. & Krystal, M. The role of template-primer interactions in cleavage and initiation by the influenza virus polymerase. J. Gen. Virol. 76, 603–611 (1995)
Rao, P., Yuan, W. & Krug, R. M. Crucial role of CA cleavage sites in the cap-snatching mechanism for initiating viral mRNA synthesis. EMBO J. 22, 1188–1198 (2003)
Geerts-Dimitriadou, C., Goldbach, R. & Kormelink, R. Preferential use of RNA leader sequences during influenza A transcription initiation in vivo. Virology 409, 27–32 (2011)
Geerts-Dimitriadou, C., Zwart, M. P., Goldbach, R. & Kormelink, R. Base-pairing promotes leader selection to prime in vitro influenza genome transcription. Virology 409, 17–26 (2011)
Braam, J., Ulmanen, I. & Krug, R. M. Molecular model of a eucaryotic transcription complex: functions and movements of influenza P proteins during capped RNA-primed transcription. Cell 34, 609–618 (1983)
Li, M. L., Rao, P. & Krug, R. M. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 20, 2078–2086 (2001)
Honda, A., Mizumoto, K. & Ishihama, A. Two separate sequences of PB2 subunit constitute the RNA cap-binding site of influenza virus RNA polymerase. Genes Cells 4, 475–485 (1999)
Shih, S. R. & Krug, R. M. Surprising function of the three influenza viral polymerase proteins: selective protection of viral mRNAs against the cap-snatching reaction catalyzed by the same polymerase proteins. Virology 226, 430–435 (1996)
Lim, K. et al. Biophysical characterization of sites of host adaptive mutation in the influenza A virus RNA polymerase PB2 RNA-binding domain. Int. J. Biochem. Cell Biol. 53, 237–245 (2014)
Ng, A. K. et al. Influenza polymerase activity correlates with the strength of interaction between nucleoprotein and PB2 through the host-specific residue K/E627. PLoS ONE 7, e36415 (2012)
Coloma, R. et al. The structure of a biologically active influenza virus ribonucleoprotein complex. PLoS Pathog. 5, e1000491 (2009)
Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D 66, 133–144 (2010)
Read, R. J. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D 57, 1373–1382 (2001)
Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D 59, 2023–2030 (2003)
Abrahams, J. P. & Leslie, A. G. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D 52, 30–42 (1996)
Murshudov, G. N. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Nicholls, R. A., Fischer, M., McNicholas, S. & Murshudov, G. N. Conformation-independent structural comparison of macromolecules with ProSMART. Acta Crystallogr. D 70, 2487–2499 (2014)
DeLano, W. L. The PyMOL Molecular Graphics System; http://www.pymol.sourceforge.net (Schrödinger, LLC, 2002)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
De la Peña, M., Kyrieleis, O. J. & Cusack, S. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. EMBO J. 26, 4913–4925 (2007)
Ye, Q., Krug, R. M. & Tao, Y. J. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature 444, 1078–1082 (2006)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comp. Chem. 25, 1605–1612 (2004)
Siebert, X. & Navaza, J. UROX 2.0: an interactive tool for fitting atomic models into electron-microscopy reconstructions. Acta Crystallogr. D 65, 651–658 (2009)
Esnouf, R. M. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D 55, 938–940 (1999)
Area, E. et al. 3D structure of the influenza virus polymerase complex: localization of subunit domains. Proc. Natl Acad. Sci. USA 101, 308–313 (2004)
Wakai, C., Iwama, M., Mizumoto, K. & Nagata, K. Recognition of cap structure by influenza B virus RNA polymerase is less dependent on the methyl residue than recognition by influenza A virus polymerase. J. Virol. 85, 7504–7512 (2011)
Acknowledgements
We thank the staff of the European Molecular Biology Laboratory (EMBL) eukaryotic expression and high-throughput crystallization facilities within the Partnership for Structural Biology (PSB) and members of the ESRF-EMBL Joint Structural Biology Group for help on European Synchrotron Radiation Facility (ESRF) beamlines. The work was supported by ERC Advanced Grant V-RNA (322586) and EU Grant FLU-PHARM (259751) to S.C. and partially by a Roche Postdoc Fellowship to S.R.
Author information
Authors and Affiliations
Contributions
S.R., D.G. and T.L. did protein expression, purification, crystallization and activity assays. A.P. did crystallographic analysis. H.M. did electron microscopy and fitting to the mini-RNP electron microscopy map. M.N. calculated the first interpretable FluB polymerase electron density map. Using the polyprotein vector designed and provided by I.B., and with the help of D.H., S.C. designed the FluB polymerase construct. T.C., D.H., R.R. and S.C. have long-collaborated on studies of influenza polymerase. S.C. supervised the project, collected data, did crystallographic analysis and wrote the paper with input from S.R., D.G., A.P., H.M. and M.N.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Production and characterization of influenza B polymerase heterotrimer.
a, Schematic of the self-cleaving polyprotein construct used to express recombinant influenza B heterotrimeric polymerase in insect cells. N-terminally it encodes the tobacco etch virus (TEV) protease that cleaves C-terminal to the amino-acid sequence ENLYFQ (in italics) and releases N-terminally His-tagged PA, PB1, C-terminally Strep-tagged PB2 and cyan fluorescent protein (CFP) for facilitated expression monitoring. Arrows indicate the N-to-C-terminal direction and the termini of each mature protein. The histidine and streptavidin tags are underlined. b, Using the PB2 C-terminal strep-tag, most contaminating proteins could be separated from the polymerase as judged by 10% SDS–PAGE followed by Coomassie blue staining. Lanes ‘M’ contain the protein markers (molecular masses indicated); ‘in’, ‘ft’ and ‘w’ denote the input, flow-through and wash of the engineered streptavidin (strep-tactin) column, respectively, and ‘elution’ indicates the re-mobilization of bound heterotrimeric polymerase by a sharp gradient of d-desthiobiotin. The three subunits, PA (85.7 kDa), PB1 (86.1 kDa) and PB2 (90.8 kDa), run together on the gel. c, After ammonium sulphate precipitation, IMAC, strep-tactin affinity and heparin chromatography, the final purification step consists of size-exclusion chromatography. The elution profile (monitored by the absorbance at 280 nm) with a single and nearly symmetric peak suggests a homogeneous and monomeric polymerase complex. d, Recombinant influenza B polymerase was analysed by electron microscopy following negative staining with sodium silico-tungstate of 0.02 mg ml−1 protein sample. The image demonstrates that the sample is homogeneous and monodisperse with a V- or doughnut-like shape with a central cavity.
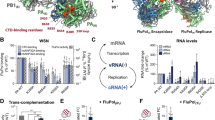
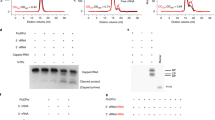
Extended Data Figure 2 Endonuclease, transcription and replication activities of FluB polymerase.
a, Schematic of mini-panhandle vRNA: 5′-pppAGUAGUAACAAGAGGGUAUUGUAUACCUCUGCUUCUGCU-3′. b, Schematic of separate 5′ and 3′ ends: 5′: 5′-pAGUAGUAACAAGAGGGUA-3′; 3′: 5′-UAUACCUCUGCUUCUGCU-3′. c, Endonuclease, cap-dependent transcription and ApG-primed replication assays. Cleavage of the cap donor is visible in lanes 2–6 and enhanced in the presence of the 5′ end, but not the 3′ end. Capped transcripts are visible in lanes 10 (from vRNA panhandle template) and 13 (from separated 5′ and 3′ vRNA ends) as well as cRNA produced in lanes 17 and 20. Markers, with size shown on the left, are RNA ladders labelled with 32P-pCp nucleotide. d, e, Time course of unprimed (d) and ApG-primed (e) vRNA replication by influenza B polymerase. The products of replication (cRNA) are indicated with an arrow. Ladders (lanes L) are 32P-pCp nucleotide-labelled RNA oligomers. ApG-primed replication is more efficient than unprimed replication.
Extended Data Figure 3 Examples of electron density map for FluB polymerase.
a–c, Initial platinum SIRAS-phased and phase-extended experimental map at 3.6 Å resolution contoured at 1.1σ (brown) with superposed final model for the FluB1 crystal form. Also shown is the final model-phased selenium anomalous difference map at 4.1 Å resolution contoured at 3.2σ (purple) highlighting methionine positions. a, PB1 β-ribbon. b, vRNA 5′ hook. c, PA–PB1–PB2 helical interface. d, e, final 2Fo − Fc omit map at 2.7 Å resolution for the FluB2 crystal form contoured at 1.1σ. d, vRNA 5′ hook nucleotides 1–11. e, vRNA 3′ end nucleotides 1–9. Figures drawn with Bobscript52.
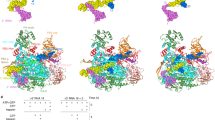
Extended Data Figure 4 Comparison of FluB and bat FluA polymerase structures.
a, Surface diagram of FluB1 structure coloured as in c except that PA-C, PB1and PB2-N are uniformly green, cyan and red, respectively. The bottom black arrow indicates the extra 12 C-terminal residues of FluB PA that extend the PA C-terminal helix compared to FluA, so that it directly contacts the PB2-NLS domain that is consequently orientated slightly differently from in FluA polymerase. b, As in a but for bat FluA structure. Arrows highlight the 70° difference in orientation of the cap-binding domain. The structural similarity between FluA and FluB polymerases (LSQMAN, cut-off 3.5 Å) is as follows. PA: 630 Cα atoms aligned, of which 38.6% are identical with root mean squared deviation (r.m.s.d.) 1.34 Å; PB1: 703 Cα atoms aligned, of which 61.3% are identical with r.m.s.d. 1.06 Å; PB2: 428 Cα atoms aligned, of which 40.6% are identical with r.m.s.d. 1.46 Å (excluding the cap-binding domain), and, taking into account the cap-binding domain rotation, 622 Cα atoms aligned, of which 39.0% are identical with r.m.s.d. 1.54 Å). c, Subunit domain structure of influenza B polymerase with names and extended colour scheme, showing the positions of the PB1 polymerase motifs. Note that for PB1, the FluB numbering compared to FluA is the same from 1–399 and is thereafter +1. For PB2, FluB is +2 from 1–469 and +1 from 470–628. For PA it is more complicated owing to several short insertions and deletions. See Supplementary Fig. 1.
Extended Data Figure 5 RNA–RNA crystal contact in FluB1 crystal form.
a, Cartoon of 5′ and 3′ vRNA ends (left, pink and yellow, respectively) interacting with crystallographic two-fold symmetry-related vRNA (right, pale pink and wheat, respectively). The PB1 β-ribbon (cyan) of the left-hand polymerase molecule interacts with the symmetry-related vRNA. b, Simplified diagram showing vRNA sequence and secondary structure in the FluB1 crystal form including vRNA-mediated crystal contact.
Extended Data Figure 6 Polymerase fitting into the mini-RNP electron microscopy map.
a, b, Top (a) and side (b) view of influenza A mini-RNP pseudo-atomic model with rescaled electron density39. PA, PB1 and PB2 (1–32 only) are shown as ribbons and coloured in green, cyan and rose, respectively. Unfilled electron density, likely to contain the rest of PB2, is shown in transparent rose. Nucleoproteins are shown in yellow ribbons, with the nucleoprotein–nucleoprotein interacting loop (residues 402–428) in orange. The vRNA 5′ and 3′ ends are shown in dark blue and red, respectively. c, Front view of influenza A mini-RNP pseudo-atomic model. The positions of antibody and tag labelling corresponding to domains of PA, PB1 and PB2 are shown as dark green, dark blue and dark rose spheres, respectively, as localized previously53. d, Close-up view of b. The PB1 β-ribbon (residues 177–214, purple) is located close to one of the proximal nucleoproteins and the vRNA. e, Putative interactions between the proximal nucleoprotein and polymerase. Nucleoprotein elements proposed for polymerase interaction are indicated in yellow, brown and orange. Polymerase interacting elements are shown in green, cyan, rose and magenta.
Extended Data Figure 7 Residual electron density in the FluB1 crystal form mimicking capped primer binding to the PB2 cap-binding domain.
Residual m2Fo − Fc (blue mesh at 0.9σ) and mFo − Fc (orange mesh at 2.5σ) electron density showing RNA-like density bound in the cap-binding site in the FluB1 crystal form. The low resolution and partial occupancy do not allow identification of the RNA and the discontinuous model shown is for illustrative purposes only. Owing to the rigorous purification procedure it is unlikely to be insect cell-capped RNA that is trapped on the polymerase. More likely it derives from the input vRNA used in crystallization, possibly partially digested by the endonuclease that generates 3′ ends. That this RNA could even be uncapped is explicable by the fact that the FluB cap-binding domain, unlike that of FluA, promiscuously binds both methylated and unmethylated guanosine54. Indeed, the density seems to be better fit with a free 3′ end sandwiched between Phe 406 and Trp 359 in the cap-binding site rather than a capped 5′ end. As the primer emerges from the cap-binding site it is initially channelled on one side by the base of the 424-loop, and on the other by residues 518–522 of the cap-627 linker. Further down, the extended 424-loop continues to guide the RNA, as well as, on the other side, the projecting N-terminal end of PB2 helix α9 (155-EMPPDE in FluB), with the double proline forcing the RNA into a ∼90° bend. Arg 425 and Arg 438 are well placed to interact with phosphates and one base seems to stack on the Glu 155–Arg 217 salt bridge. Conserved basic residues on PB2 N2 domain strands β7, 144-Arg-Lys-Arg (FluA 142-Arg-Lys-Arg), and β8, 216-Arg-Arg-Arg-Phe (FluA 214-Arg-Thr-Arg-Phe), are also likely to be involved. Straight-line distances from the cap-binding site to the bend and from the bend to the PB1 active site are indicated. See also Fig. 5.
Extended Data Figure 8 Schematic diagram of steps in cap-dependent transcription by influenza virus polymerase.
a, Cap-snatching from host pre-mRNA (red). The m7G cap is bound by the cap-binding domain (orange, orientated as in the FluA structure) and the pre-mRNA cleaved 10–14 nucleotides downstream by the endonuclease (green). The single-stranded vRNA genome is bound by its 5′ (hook, pink) and 3′ (template, yellow) ends to the polymerase (blue, depicted as a cutaway section). b, Transcription initiation. The cap-binding domain rotates to the position observed in the FluB1 structure directing the capped primer into the PB1 active site, where it potentially makes limited base pairs with the extremity of the template. Template-directed NTP addition (white) extends the host sequences (red) with virally encoded sequences (cyan). Note that in b–d additional conformational changes in the polymerase are expected, but not depicted since they are currently unknown. c, Transcription elongation. Transcription elongation proceeds, eventually leading to the release of the cap from the cap-binding domain (d) and the binding of host mRNP factors. d, Polyadenylation by stuttering. After most of the vRNA template has been translocated through the polymerase, only a tight turn connects it to the bound 5′-hook. The nucleotide sequence of this region is given at the bottom. This places the 5′ proximal oligo-U stretch in the PB1 active site allowing poly(A) tail synthesis by a stuttering mechanism in which the template is no longer translocated but the product strand is able to slip.
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1, Supplementary Text and Supplementary References. (PDF 2312 kb)
Simulation of the putative rotation of the cap-binding domain (orange) by morphing between the domain positions seen in the FluA and FluB crystal structures.
The yellow spheres in the cap-binding domain correspond to the bound cap-analogue m7GTP. (MP4 557 kb)
Rights and permissions
About this article
Cite this article
Reich, S., Guilligay, D., Pflug, A. et al. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 516, 361–366 (2014). https://doi.org/10.1038/nature14009
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14009
This article is cited by
-
Mechanisms and consequences of mRNA destabilization during viral infections
Virology Journal (2024)
-
The ubiquitination landscape of the influenza A virus polymerase
Nature Communications (2023)
-
Protein purification strategies must consider downstream applications and individual biological characteristics
Microbial Cell Factories (2022)
-
Cap-snatching inhibitors of influenza virus are inhibitory to the in vitro transcription of rice stripe virus
Phytopathology Research (2022)
-
Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.