Abstract
The influenza virus polymerase transcribes or replicates the segmented RNA genome (viral RNA) into viral messenger RNA or full-length copies. To initiate RNA synthesis, the polymerase binds to the conserved 3′ and 5′ extremities of the viral RNA. Here we present the crystal structure of the heterotrimeric bat influenza A polymerase, comprising subunits PA, PB1 and PB2, bound to its viral RNA promoter. PB1 contains a canonical RNA polymerase fold that is stabilized by large interfaces with PA and PB2. The PA endonuclease and the PB2 cap-binding domain, involved in transcription by cap-snatching, form protrusions facing each other across a solvent channel. The 5′ extremity of the promoter folds into a compact hook that is bound in a pocket formed by PB1 and PA close to the polymerase active site. This structure lays the basis for an atomic-level mechanistic understanding of the many functions of influenza polymerase, and opens new opportunities for anti-influenza drug design.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Resa-Infante, P., Jorba, N., Coloma, R. & Ortin, J. The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol. 8, 207–215 (2011)
Fodor, E. The RNA polymerase of influenza a virus: mechanisms of viral transcription and replication. Acta Virol. 57, 113–122 (2013)
Ruigrok, R. W., Crepin, T., Hart, D. J. & Cusack, S. Towards an atomic resolution understanding of the influenza virus replication machinery. Curr. Opin. Struct. Biol. 20, 104–113 (2010)
Plotch, S. J., Bouloy, M., Ulmanen, I. & Krug, R. M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23, 847–858 (1981)
Dias, A. et al. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458, 914–918 (2009)
Yuan, P. et al. Crystal structure of an avian influenza polymerase PAN reveals an endonuclease active site. Nature 458, 909–913 (2009)
Guilligay, D. et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nature Struct. Mol. Biol. 15, 500–506 (2008)
Kowalinski, E. et al. Structural analysis of specific metal chelating inhibitor binding to the endonuclease domain of influenza pH1N1 (2009) polymerase. PLoS Pathog. 8, e1002831 (2012)
Clark, M. P. et al. Discovery of a novel, first-in-class, orally bioavailable azaindole inhibitor (VX-787) of influenza PB2. J. Med. Chem. 57, 6668–6678 (2014)
He, X. et al. Crystal structure of the polymerase PAC–PB1N complex from an avian influenza H5N1 virus. Nature 454, 1123–1126 (2008)
Obayashi, E. et al. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454, 1127–1131 (2008)
Sugiyama, K. et al. Structural insight into the essential PB1–PB2 subunit contact of the influenza virus RNA polymerase. EMBO J. 28, 1803–1811 (2009)
Tarendeau, F. et al. Host determinant residue lysine 627 lies on the surface of a discrete, folded domain of influenza virus polymerase PB2 subunit. PLoS Pathog. 4, e1000136 (2008)
Cauldwell, A. V., Long, J. S., Moncorge, O. & Barclay, W. S. Viral determinants of influenza A virus host range. J. Gen. Virol. 95, 1193–1210 (2014)
Tarendeau, F. et al. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nature Struct. Mol. Biol. 14, 229–233 (2007)
Tong, S. et al. A distinct lineage of influenza A virus from bats. Proc. Natl Acad. Sci. USA 109, 4269–4274 (2012)
Poole, D. S. et al. Influenza A virus polymerase is a site for adaptive changes during experimental evolution in bat cells. J. Virol. 88, 12572–12585 (2014)
Reich, S. et al. Structural insights into cap-snatching and RNA synthesis by influenza virus polymerase. Nature http://dx.doi.org/10.1038/nature14009 (this issue)
Turrell, L., Lyall, J. W., Tiley, L. S., Fodor, E. & Vreede, F. T. The role and assembly mechanism of nucleoprotein in influenza A virus ribonucleoprotein complexes. Nature Commun. 4, 1591 (2013)
González, S., Zurcher, T. & Ortin, J. Identification of two separate domains in the influenza virus PB1 protein involved in the interaction with the PB2 and PA subunits: a model for the viral RNA polymerase structure. Nucleic Acids Res. 24, 4456–4463 (1996)
Poole, E. L., Medcalf, L., Elton, D. & Digard, P. Evidence that the C-terminal PB2-binding region of the influenza A virus PB1 protein is a discrete α-helical domain. FEBS Lett. 581, 5300–5306 (2007)
Müller, R., Poch, O., Delarue, M., Bishop, D. H. & Bouloy, M. Rift Valley fever virus L segment: correction of the sequence and possible functional role of newly identified regions conserved in RNA-dependent polymerases. J. Gen. Virol. 75, 1345–1352 (1994)
Biswas, S. K. & Nayak, D. P. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J. Virol. 68, 1819–1826 (1994)
Bruenn, J. A. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res. 31, 1821–1829 (2003)
Yap, T. L. et al. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J. Virol. 81, 4753–4765 (2007)
Lesburg, C. A. et al. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nature Struct. Biol. 6, 937–943 (1999)
Bressanelli, S. et al. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl Acad. Sci. USA 96, 13034–13039 (1999)
Butcher, S. J., Grimes, J. M., Makeyev, E. V., Bamford, D. H. & Stuart, D. I. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410, 235–240 (2001)
Caillet-Saguy, C., Lim, S. P., Shi, P. Y., Lescar, J. & Bressanelli, S. Polymerases of hepatitis C viruses and flaviviruses: structural and mechanistic insights and drug development. Antiviral Res. 105, 8–16 (2014)
Hutchinson, E. C., Orr, O. E., Man Liu, S., Engelhardt, O. G. & Fodor, E. Characterization of the interaction between the influenza A virus polymerase subunit PB1 and the host nuclear import factor Ran-binding protein 5. J. Gen. Virol. 92, 1859–1869 (2011)
Kuzuhara, T. et al. Structural basis of the influenza A virus RNA polymerase PB2 RNA-binding domain containing the pathogenicity-determinant lysine 627 residue. J. Biol. Chem. 284, 6855–6860 (2009)
Pautus, S. et al. New 7-methylguanine derivatives targeting the influenza polymerase PB2 cap-binding domain. J. Med. Chem. 56, 8915–8930 (2013)
Zamyatkin, D. F. et al. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J. Biol. Chem. 283, 7705–7712 (2008)
Fodor, E., Pritlove, D. C. & Brownlee, G. G. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 68, 4092–4096 (1994)
Tiley, L. S., Hagen, M., Matthews, J. T. & Krystal, M. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J. Virol. 68, 5108–5116 (1994)
Hsu, M. T., Parvin, J. D., Gupta, S., Krystal, M. & Palese, P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc. Natl Acad. Sci. USA 84, 8140–8144 (1987)
Neumann, G. & Hobom, G. Mutational analysis of influenza virus promoter elements in vivo. J. Gen. Virol. 76, 1709–1717 (1995)
Flick, R., Neumann, G., Hoffmann, E., Neumeier, E. & Hobom, G. Promoter elements in the influenza vRNA terminal structure. RNA 2, 1046–1057 (1996)
Fodor, E., Pritlove, D. C. & Brownlee, G. G. Characterization of the RNA-fork model of virion RNA in the initiation of transcription in influenza A virus. J. Virol. 69, 4012–4019 (1995)
Kim, H. J., Fodor, E., Brownlee, G. G. & Seong, B. L. Mutational analysis of the RNA-fork model of the influenza A virus vRNA promoter in vivo. J. Gen. Virol. 78, 353–357 (1997)
Pritlove, D. C., Poon, L. L., Devenish, L. J., Leahy, M. B. & Brownlee, G. G. A hairpin loop at the 5′ end of influenza A virus virion RNA is required for synthesis of poly(A)+ mRNA in vitro. J. Virol. 73, 2109–2114 (1999)
Briese, T. et al. Upolu virus and Aransas Bay virus, two presumptive bunyaviruses, are novel members of the family Orthomyxoviridae. J. Virol. 88, 5298–5309 (2014)
Fodor, E. et al. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76, 8989–9001 (2002)
González, S. & Ortin, J. Characterization of influenza virus PB1 protein binding to viral RNA: two separate regions of the protein contribute to the interaction domain. J. Virol. 73, 631–637 (1999)
Li, M. L., Ramirez, B. C. & Krug, R. M. RNA-dependent activation of primer RNA production by influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J. 17, 5844–5852 (1998)
Jung, T. E. & Brownlee, G. G. A new promoter-binding site in the PB1 subunit of the influenza A virus polymerase. J. Gen. Virol. 87, 679–688 (2006)
Kerry, P. S., Willsher, N. & Fodor, E. A cluster of conserved basic amino acids near the C-terminus of the PB1 subunit of the influenza virus RNA polymerase is involved in the regulation of viral transcription. Virology 373, 202–210 (2008)
Leahy, M. B., Pritlove, D. C., Poon, L. L. & Brownlee, G. G. Mutagenic analysis of the 5′ arm of the influenza A virus virion RNA promoter defines the sequence requirements for endonuclease activity. J. Virol. 75, 134–142 (2001)
Rao, P., Yuan, W. & Krug, R. M. Crucial role of CA cleavage sites in the cap-snatching mechanism for initiating viral mRNA synthesis. EMBO J. 22, 1188–1198 (2003)
Poon, L. L., Pritlove, D. C., Sharps, J. & Brownlee, G. G. The RNA polymerase of influenza virus, bound to the 5′ end of virion RNA, acts in cis to polyadenylate mRNA. J. Virol. 72, 8214–8219 (1998)
Nie, Y., Bellon-Echeverria, I., Trowitzsch, S., Bieniossek, C. & Berger, I. Multiprotein complex production in insect cells by using polyproteins. Methods Mol. Biol. 1091, 131–141 (2014)
Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D 66, 133–144 (2010)
Tefsen, B. et al. The N-terminal domain of PA from bat-derived influenza-like virus H17N10 has endonuclease activity. J. Virol. 88, 1935–1941 (2014)
Murshudov, G. N. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
DeLano, W. L. The PyMOL Molecular Graphics System; http://www.pymol.sourceforge.net (Schrödinger, LLC, 2002)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. 66, 12–21 (2010)
De la Peña, M., Kyrieleis, O. J. & Cusack, S. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. EMBO J. 26, 4913–4925 (2007)
Acknowledgements
We thank members of the ESRF-EMBL Joint Structural Biology Group for access to European Synchrotron Radiation Facility (ESRF) beamlines, staff of the European Molecular Biology Laboratory (EMBL) eukaryotic expression and high-throughput crystallization facilities within the Partnership for Structural Biology (PSB), D. Hart for help with construct design, and H. Malet for electron microscopy. This work was supported by ERC Advanced Grant V-RNA (322586) to S.C.
Author information
Authors and Affiliations
Contributions
A.P. did protein expression, purification and crystallization, with help from S.R. and D.G. who also did activity assays. A.P. did X-ray data collection and, together with S.C., did crystallographic analysis. S.C. supervised the project and wrote the paper with input from the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Production of influenza A polymerase heterotrimer.
a, The heterotrimeric bat polymerase was recombinantly expressed in insect cells as a self-cleaving polyprotein. N-terminally it encodes the tobacco etch virus (TEV) protease that cleaves C-terminal to the amino-acid sequence ENLYFQ (in italics), and releases N-terminally His-tagged PA, PB1, C-terminally strep-tagged PB2 and cyan fluorescent protein (CFP) for facilitated monitoring of expression. Arrows indicate the N-to-C-terminal direction and the termini of each mature protein. The histidine and streptavidin tags are underlined. b, After ammonium sulphate precipitation, immobilized metal ion affinity chromatography, engineered streptavidin (strep-tactin) affinity and heparin chromatography, the final purification step consisted of size-exclusion chromatography. The elution profile (monitored by the absorbance at 280 nm) with a single and nearly symmetric peak suggests a homogeneous and monomeric polymerase complex. mAU, milli-absorption unit. c, Fractions of the final size-exclusion chromatography were subjected to 10% SDS–PAGE followed by Coomassie blue staining. Lane 1 contains the molecular mass markers and lanes 2–7 the eluate with PA (85.4 kilodaltons (kDa)), PB1 (87.8 kDa) and PB2 (91.0 kDa). d, Recombinant bat FluA polymerase was visualized by electron microscopy following negative staining with sodium silico-tungstate of a 0.02 mg ml−1 protein sample. The image demonstrates that the sample is homogeneous and monodisperse with a V- or doughnut-like shape and central cavity.
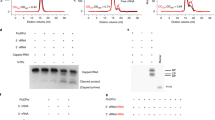
Extended Data Figure 2 Endonuclease, RNA transcription and RNA replication activities of recombinant FluA polymerase.
a, Mini-panhandle vRNA: 5′-pppAGUAGUAACAAGAGGGUAUUGUAUACCUCUGCUUCUGCU-3′. b, Separate 5′ and 3′ ends: 5′: 5′-pAGUAGUAACAAGAGGGUA-3′; 3′: 5′-UAUACCUCUGCUUCUGCU-3′. c, Endonuclease, cap-dependent transcription and ApG-primed replication assays. Cleavage of the cap donor is visible in lanes 2–6. Capped transcripts are visible in lanes 10 (from vRNA panhandle template) and 13 (from separated 5′ and 3′ vRNA ends) as well as cRNA produced in lanes 17 and 20. Markers, with size shown on the left, are RNA ladders labelled with 32P-pCp nucleotide. d, e, Time course of unprimed (d) and ApG-primed (e) vRNA replication by bat influenza A polymerase. The products of replication (cRNA) are indicated with an arrow. Ladders (lanes L) are 32P-pCp nucleotide-labelled RNA oligomers. ApG-primed replication is more efficient than unprimed replication.
Extended Data Figure 3 Surface views of the FluA heterotrimer with bound vRNA promoter.
a–d, Four surface views at roughly 0° (a), 180° (b), 110° (c) and 290° (d) rotations with PA, PB1 and PB2 uniformly green, cyan and red, respectively. Major subdomains are labelled. The vRNA 5′ and 3′ ends are pink and yellow, respectively.
Extended Data Figure 4 PA and PB2 structure and new inter-subunit interactions.
a, Interactions of the PA-linker (green tube) with the outer surface of the fingers (pale cyan) and palm (pale salmon) domains of PB1. Contacts are mediated by both highly conserved hydrophobic residues (for example, PA residues Phe 205, Phe 211, Leu 214, Pro 220, Tyr 226, Phe 229, Tyr 232, Val 233, Ile 242, Leu 246, Met 249 and Val 253) and polar interactions (for example, PA Glu 203, Lys 230, Glu 243 and Lys 245 to PB1 Arg 162, Glu 331, His 465 and Asp 86, respectively). b, Transparent surface diagram showing the anchoring of the PA endonuclease domain (forest green) onto the PB1-Cter–PB2-Nter interface region (cyan/red) and its position relative to the PB2 cap-binding domain (orange). The nuclease helix α4 packs parallel to the penultimate PB1 helix α21 involving both hydrophobic (for example, PA Ile 86, Ile 90 and Ile 94 with PB1 Ser 720, Ile 724 and Ile 728, respectively) and polar interactions (for example, PA Glu 77 with PB1 Arg 727). Other contacts include the PB2 170-loop interacting with the same PA helix α4 in the vicinity of Trp 88. Also the endonuclease insertion (PA 70-loop, residues 67–74) packs on the first part of the last PB1 helix α22. The total buried surface area between the endonuclease and PB1/PB2 is 2,265 Å2.
Extended Data Figure 5 NTP and template tunnels in PB1.
a, View straight along the putative NTP entrance tunnel towards the putative priming loop (magenta) in the internal cavity. The NTP channel is lined with basic residues from the fingertips (Lys 235, Lys 237 and Arg 239, blue), fingers (Arg 45, cyan) and palm (Lys 308, Lys 480 and Lys 481, red) that are absolutely conserved in all influenza strains. The fingertips are in close proximity to PA helices α20 and α21 and to the loop of the 5′ hook. b, Surface view as in a showing that the putative priming loop in the interior cavity is visible through the NTP tunnel. c, View straight along the template entrance tunnel towards the priming loop (magenta) in the internal cavity. The tunnel is lined by residues conserved in all influenza strains and from all three subunits, Arg 507 and Asp 509 from PA (green), Tyr 30, Arg 126, Met 227, Lys 229 and Asp 230 from PB1 (cyan), and Arg 38, Lys 41 and Asn 42 from PB2 (red). d, Surface view as in c showing that the internal priming loop is visible through the template tunnel.
Extended Data Figure 6 Recognition of the vRNA 3′ end.
Protein interactions of the distal 3′ end showing the role of PB2-Nter (red). PB2 residues Arg 46 and Trp 49 and PA residue Lys 567 stabilize the sharp turn between 3′ nucleotides C8 and G9. PB2 Arg 38 and PB1-Cter residues Asn 671, Arg 672 and Asn 676 also bind the 3′ end. In the accompanying paper18, Fig. 2a shows the interactions with the complete 3′ end as observed in the FluB vRNA complex.
Extended Data Figure 7 vRNA arrangement in the bat polymerase crystals.
Simplified diagram showing vRNA sequence and secondary structure in the bat FluA crystals including vRNA-mediated crystal contact (inverted sequences) that forms an extended duplex. Crystals were grown with 3′-end nucleotides 1–18 or 3–18, but only those from 6–18 were visible (hence 1–5 are in italics).
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1, Supplementary Discussions and Supplementary References. (PDF 1975 kb)
360° rotation of bat FluA structure in ribbon representation about the vertical axis
View and colouring as in Fig. 1 (AVI 16280 kb)
360° rotation of bat FluA structure in surface representation about the vertical axis
View and colouring as in Fig. 1. (AVI 24293 kb)
Rights and permissions
About this article
Cite this article
Pflug, A., Guilligay, D., Reich, S. et al. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516, 355–360 (2014). https://doi.org/10.1038/nature14008
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14008
This article is cited by
-
The ubiquitination landscape of the influenza A virus polymerase
Nature Communications (2023)
-
Phosphorylation of the PA subunit of influenza polymerase at Y393 prevents binding of the 5′-termini of RNA and polymerase function
Scientific Reports (2023)
-
ERDRP-0519 inhibits feline coronavirus in vitro
BMC Veterinary Research (2022)
-
Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies
Nature Communications (2022)
-
Structure of Machupo virus polymerase in complex with matrix protein Z
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.