Abstract
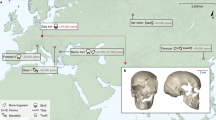
We present the high-quality genome sequence of a ∼45,000-year-old modern human male from Siberia. This individual derives from a population that lived before—or simultaneously with—the separation of the populations in western and eastern Eurasia and carries a similar amount of Neanderthal ancestry as present-day Eurasians. However, the genomic segments of Neanderthal ancestry are substantially longer than those observed in present-day individuals, indicating that Neanderthal gene flow into the ancestors of this individual occurred 7,000–13,000 years before he lived. We estimate an autosomal mutation rate of 0.4 × 10−9 to 0.6 × 10−9 per site per year, a Y chromosomal mutation rate of 0.7 × 10−9 to 0.9 × 10−9 per site per year based on the additional substitutions that have occurred in present-day non-Africans compared to this genome, and a mitochondrial mutation rate of 1.8 × 10−8 to 3.2 × 10−8 per site per year based on the age of the bone.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Primary accessions
European Nucleotide Archive
Data deposits
All sequence data have been submitted to the European Nucleotide Archive (ENA) and are available under the following Ust’-Ishim accession number: PRJEB6622. The data from the 25 present-day human genomes are available from (http://www.simonsfoundation.org/life-sciences/simons-genome-diversity-project/) and from (http://cdna.eva.mpg.de/neandertal/altai/).
References
Trinkaus, E. & Ruff, C. B. Diaphyseal cross-sectional geometry of Near Eastern Middle Paleolithic humans: the femur. J. Archaeol. Sci. 26, 409–424 (1999)
Brock, F. et al. Reliability of nitrogen content (%N) and carbon:nitrogen atomic ratios (C:N) as indicators of collagen preservation suitable for radiocarbon dating. Radiocarbon 54, 879–886 (2012)
Richards, M. P. & Trinkaus, E. Out of Africa: modern human origins special feature: isotopic evidence for the diets of European Neanderthals and early modern humans. Proc. Natl Acad. Sci. USA 106, 16034–16039 (2009)
Meyer, M. et al. A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012)
Fu, Q. et al. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr. Biol. 23, 553–559 (2013)
The Y Chromosome Consortium A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 12, 339–348 (2002)
Shapiro, B. et al. A Bayesian phylogenetic method to estimate unknown sequence ages. Mol. Biol. Evol. 28, 879–887 (2011)
Patterson, N. et al. Ancient admixture in human history. Genetics 192, 1065–1093 (2012)
Olalde, I. et al. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature 507, 225–228 (2014)
Raghavan, M. et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 505, 87–91 (2014)
Lazaridis, I. et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014)
Prüfer, K. et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014)
Li, H. & Durbin, R. Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011)
Scally, A. & Durbin, R. Revising the human mutation rate: implications for understanding human evolution. Nature Rev. Genet. 13, 745–753 (2012)
Kong, A. et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488, 471–475 (2012)
Langergraber, K. E. et al. Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc. Natl Acad. Sci. USA 109, 15716–15721 (2012)
Prüfer, K. et al. The bonobo genome compared with the chimpanzee and human genomes. Nature 486, 527–531 (2012)
Xue, Y. et al. Human Y chromosome base-substitution mutation rate measured by direct sequencing in a deep-rooting pedigree. Curr. Biol. 19, 1453–1457 (2009)
Kuroki, Y. et al. Comparative analysis of chimpanzee and human Y chromosomes unveils complex evolutionary pathway. Nature Genet. 38, 158–167 (2006)
Wang, J., Fan, H. C., Behr, B. & Quake, S. R. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell 150, 402–412 (2012)
Sankararaman, S., Patterson, N., Li, H., Pääbo, S. & Reich, D. The date of interbreeding between Neandertals and modern humans. PLoS Genet. 8, e1002947 (2012)
Reich, D. et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060 (2010)
Reich, D. et al. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am. J. Hum. Genet. 89, 516–528 (2011)
Skoglund, P. & Jakobsson, M. Archaic human ancestry in East Asia. Proc. Natl Acad. Sci. USA 108, 18301–18306 (2011)
Fenner, J. N. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am. J. Phys. Anthropol. 128, 415–423 (2005)
McCown, T. D. & Keith, A. The Stone Age of Mount Carmel Vol. 2 (Clarendon, Oxford, 1939)
Vandermeersch, B. Les Hommes Fossiles de Qafzeh (Israel) 319 (Éditions du CNRS, 1981)
Rasmussen, M. et al. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science 334, 94–98 (2011)
Hublin, J. J. The earliest modern human colonization of Europe. Proc. Natl Acad. Sci. USA 109, 13471–13472 (2012)
Müller, U. C. et al. The role of climate in the spread of modern humans into Europe. Quat. Sci. Rev. 30, 273–279 (2011)
Goebel, T. A., Derevianko, A. P. & Petrin, V. T. Dating the Middle to Upper Paleolithic transition at Kara-Bom. Curr. Anthropol. 34, 452–458 (1993)
Kuhn, S. L. & Zwyns, N. Rethinking the initial Upper Paleolithic. Quat. Int. http://dx.doi.org/10.1016/j.quaint.2014.05.040 (2014)
Bronk Ramsey, C., Scott, M. & van der Plicht, H. Calibration for archaeological and environmental terrestrial samples in the time range 26–50 ka cal bp. Radiocarbon. 55, 2021–2027 (2013)
Reimer, P. J. et al. IntCal13 and Marine13 radiocarbon age calibration curves 0–50,000 Years cal bp. Radiocarbon 55, 1869–1887 (2009)
Kircher, M., Stenzel, U. & Kelso, J. Improved base calling for the Illumina Genome Analyzer using machine learning strategies. Genome Biol. 10, R83 (2009)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
Acknowledgements
We are grateful to P. Gunz, M. Kircher, A. I. Krivoshapkin, P. Nigst, M. Ongyerth, N. Patterson, G. Renaud, U. Stenzel, M. Stoneking and S. Talamo for valuable input, comments and help; T. Pfisterer and H. Temming for technical assistance. Q.F. is funded in part by the Chinese Academy of Sciences (XDA05130202) and the Ministry of Science and Technology of China (2007FY110200); P.A.K. by Urals Branch, Russian Academy of Sciences (12-C-4-1014) and Y.V.K. by the Russian Foundation for Basic Sciences (12-06-00045); F.J. and M.S. by the National Institutes of Health of the USA (R01-GM40282); P.J. by the NIH (K99-GM104158); and T.F.G.H. by ERC advanced grant 324139. D.R. is a Howard Hughes Medical Institute Investigator and supported by the National Science Foundation (1032255) and the NIH (GM100233). Major funding for this work was provided by the Presidential Innovation Fund of the Max Planck Society.
Author information
Authors and Affiliations
Contributions
Q.F., S.M.S., A.A.B., Y.V.K., J.K., T.B.V. and S.P. designed the research. A.A.P. and Q.F. performed the experiments; Q.F., H.L., P.M., F.J., P.L.F.J., K.P., C.d.F., M.M., M.L., M.S., D.R., J.K. and S.P. analysed genetic data; K.D. and T.F.G.H. performed 14C dating; D.C.S.-G. and M.P.R. analysed stable isotope data; N.V.P., P.A.K. and D.I.R. contributed samples and data; S.M.S., A.A.B., N.Z., Y.V.K., S.G.K., J.-J.H. and T.B.V. analysed archaeological and anthropological data; Q.F., J.K., T.B.V. and S.P. wrote and edited the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Information Sections 1-18 – see Supplementary Contents for details (PDF 25041 kb)
Rights and permissions
About this article
Cite this article
Fu, Q., Li, H., Moorjani, P. et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514, 445–449 (2014). https://doi.org/10.1038/nature13810
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13810
This article is cited by
-
The Allen Ancient DNA Resource (AADR) a curated compendium of ancient human genomes
Scientific Data (2024)
-
More than a decade of genetic research on the Denisovans
Nature Reviews Genetics (2024)
-
Delayed increase in stone tool cutting-edge productivity at the Middle-Upper Paleolithic transition in southern Jordan
Nature Communications (2024)
-
New dating indicates intermittent human occupation of the Nwya Devu Paleolithic site on the high-altitude central Tibetan Plateau during the past 45,000 years
Science China Earth Sciences (2024)
-
Us and Them: How to Reconcile Archaeological and Biological Data at the Middle-to-Upper Palaeolithic Transition in Europe?
Journal of Paleolithic Archaeology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.