Abstract
The CRISPR-associated protein Cas9 is an RNA-guided DNA endonuclease that uses RNA–DNA complementarity to identify target sites for sequence-specific double-stranded DNA (dsDNA) cleavage1,2,3,4,5. In its native context, Cas9 acts on DNA substrates exclusively because both binding and catalysis require recognition of a short DNA sequence, known as the protospacer adjacent motif (PAM), next to and on the strand opposite the twenty-nucleotide target site in dsDNA4,5,6,7. Cas9 has proven to be a versatile tool for genome engineering and gene regulation in a large range of prokaryotic and eukaryotic cell types, and in whole organisms8, but it has been thought to be incapable of targeting RNA5. Here we show that Cas9 binds with high affinity to single-stranded RNA (ssRNA) targets matching the Cas9-associated guide RNA sequence when the PAM is presented in trans as a separate DNA oligonucleotide. Furthermore, PAM-presenting oligonucleotides (PAMmers) stimulate site-specific endonucleolytic cleavage of ssRNA targets, similar to PAM-mediated stimulation of Cas9-catalysed DNA cleavage7. Using specially designed PAMmers, Cas9 can be specifically directed to bind or cut RNA targets while avoiding corresponding DNA sequences, and we demonstrate that this strategy enables the isolation of a specific endogenous messenger RNA from cells. These results reveal a fundamental connection between PAM binding and substrate selection by Cas9, and highlight the utility of Cas9 for programmable transcript recognition without the need for tags.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Wiedenheft, B., Sternberg, S. H. & Doudna, J. A. RNA-guided genetic silencing systems in bacteria and archaea. Nature 482, 331–338 (2012)
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007)
Garneau, J. E. et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 (2010)
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012)
Gasiunas, G., Barrangou, R., Horvath, P. & Siksnys, V. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl Acad. Sci. USA 109, E2579–E2586 (2012)
Mojica, F. J. M., Diez-Villasenor, C., Garcia-Martinez, J. & Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiol 155, 733–740 (2009)
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C. & Doudna, J. A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67 (2014)
Mali, P., Esvelt, K. M. & Church, G. M. Cas9 as a versatile tool for engineering biology. Nature Methods 10, 957–963 (2013)
Marraffini, L. A. & Sontheimer, E. J. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571 (2010)
Sashital, D. G., Wiedenheft, B. & Doudna, J. A. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol. Cell 46, 606–615 (2012)
Pommer, A. J. et al. Mechanism and cleavage specificity of the H-N-H endonuclease colicin E9. J. Mol. Biol. 314, 735–749 (2001)
Hsia, K. C. et al. DNA binding and degradation by the HNH protein ColE7. Structure 12, 205–214 (2004)
Nishimasu, H. et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949 (2014)
Wu, H. J., Lima, W. F. & Crooke, S. T. Properties of cloned and expressed human RNase H1. J. Biol. Chem. 274, 28270–28278 (1999)
Engreitz, J. M. et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, (2013)
Chu, C., Qu, K., Zhong, F. L., Artandi, S. E. & Chang, H. Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA–chromatin interactions. Mol. Cell 44, 667–678 (2011)
Simon, M. D. et al. The genomic binding sites of a noncoding RNA. Proc. Natl Acad. Sci. USA 108, 20497–20502 (2011)
Mackay, J. P., Font, J. & Segal, D. J. The prospects for designer single-stranded RNA-binding proteins. Nature Struct. Mol. Biol. 18, 256–261 (2011)
Filipovska, A. & Rackham, O. Designer RNA-binding proteins: new tools for manipulating the transcriptome. RNA Biol. 8, 978–983 (2011)
Wang, Y., Wang, Z. & Tanaka Hall, T. M. Engineered proteins with Pumilio/fem-3 mRNA binding factor scaffold to manipulate RNA metabolism. FEBS J. 280, 3755–3767 (2013)
Yin, P. et al. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504, 168–171 (2013)
Yagi, Y., Nakamura, T. & Small, I. The potential for manipulating RNA with pentatricopeptide repeat proteins. Plant J. 78, 772–782 (2014)
Kim, D. H. & Rossi, J. J. RNAi mechanisms and applications. Biotechniques 44, 613–616 (2008)
Hale, C. R. et al. RNA-guided RNA cleavage by a CRISPR RNA–Cas protein complex. Cell 139, 945–956 (2009)
Hale, C. R. et al. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell 45, 292–302 (2012)
Staals, R. H. J. et al. Structure and activity of the RNA-targeting type III-B CRISPR-Cas complex of Thermus thermophilus. Mol. Cell 52, 135–145 (2013)
Spilman, M. et al. Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol. Cell 52, 146–152 (2013)
Terns, R. M. & Terns, M. P. CRISPR-based technologies: prokaryotic defense weapons repurposed. Trends Genet. 30, 111–118 (2014)
Sampson, T. R., Saroj, S. D., Llewellyn, A. C., Tzeng, Y. L. & Weiss, D. S. A. CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 497, 254–257 (2013)
Sampson, T. R. & Weiss, D. S. Exploiting CRISPR/Cas systems for biotechnology. Bioessays 36, 34–38 (2014)
Sternberg, S. H., Haurwitz, R. E. & Doudna, J. A. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA 18, 661–672 (2012)
Chen, B. et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155, 1479–1491 (2013)
Lee, H. Y. et al. RNA-protein analysis using a conditional CRISPR nuclease. Proc. Natl Acad. Sci. USA 110, 5416–5421 (2013)
Chomczynski, P. One-hour downward alkaline capillary transfer for blotting of DNA and RNA. Anal. Biochem. 201, 134–139 (1992)
Acknowledgements
We thank B. Staahl and K. Zhou for technical assistance, A. Iavarone for assistance with mass spectrometry measurements, Integrated DNA Technologies for the synthesis of DNA and RNA oligonucleotides, and members of the Doudna laboratory and J. Cate for discussions and critical reading of the manuscript. S.H.S. acknowledges support from the National Science Foundation and National Defense Science & Engineering Graduate Research Fellowship programs. A.E.-S. and B.L.O. acknowledge support from NIH NRSA trainee grants. Funding was provided by the NIH-funded Center for RNA Systems Biology (P50GM102706-03). J.A.D. is an Investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
M.R.O. and S.H.S. conceived the project. M.R.O., B.L.O., S.H.S., A.E.-S. and M.K. conducted experiments. All authors discussed the data, and M.R.O., S.H.S., B.L.O. and J.A.D. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.A.D., M.R.O., B.L.O. and S.H.S. are inventors on a related patent application.
Extended data figures and tables
Extended Data Figure 1 Quantified data for cleavage of ssRNA by Cas9–gRNA in the presence of a 19-nucleotide PAMmer.
Cleavage assays were conducted as described in the Methods, and the quantified data were fitted with single-exponential decays. Results from four independent experiments yielded an average apparent pseudo-first-order cleavage rate constant (mean ± s.d.) of 0.032 ± 0.007 min−1. This is slower than the rate constant determined previously for ssDNA in the presence of the same 19-nucleotide PAMmer (7.3 ± 3.2 min−1)7.
Extended Data Figure 2 RNA cleavage is marginally stimulated by di- and tri-deoxyribonucleotide PAMmers.
Cleavage reactions contained ∼1 nM 5′-radiolabelled target ssRNA and no PAMmer (left), 100 nM 18-nt PAMmer (second from left), or 1 mM of the indicated di- or tri-nucleotide (remaining lanes). Reaction products were resolved by 12% denaturing polyacrylamide gel electrophoresis (PAGE) and visualized by phosphorimaging.
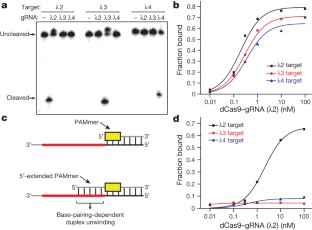
Extended Data Figure 3 Representative binding experiment demonstrating guide-specific ssRNA binding with 5′-extended PAMmers.
Gel shift assays were conducted as described in the Methods. Binding reactions contained Cas9 programmed with λ2 gRNA and either λ2 (on-target), λ3 (off-target) or λ4 (off-target) ssRNA in the presence of short cognate PAMmers or cognate PAMmers with complete 5′-extensions, as indicated. The presence of a cognate 5′-extended PAM-mer abrogates off-target binding. Three independent experiments were conducted to produce the data shown in Fig. 3b, d.
Extended Data Figure 4 Exploration of RNA cleavage efficiencies and binding specificity using PAMmers with variable 5′-extensions.
a, Cleavage assays were conducted as described in Methods. Reactions contained Cas9 programmed with λ2 gRNA and λ2 ssRNA targets in the presence of PAMmers with 5′-extensions of variable length. The ssRNA cleavage efficiency decreases as the PAMmer extends further into the target region, as indicated by the fraction of RNA cleaved after 1 h. b, Binding assays were conducted as described in the Methods, using mostly the same panel of 5′-extended PAMmers as in a. Binding reactions contained Cas9 programmed with λ2 gRNA and either λ2 (on-target) or λ3 (off-target) ssRNA in the presence of cognate PAMmers with 5′-extensions of variable length. The binding specificity increases as the PAMmer extends further into the target region, as indicated by the fraction of λ3 (off-target) ssRNA bound at 3 nM Cas9–gRNA. PAMmers with 5′ extensions also cause a slight reduction in the relative binding affinity of λ2 (on-target) ssRNA.
Extended Data Figure 5 Site-specific biotin labelling of Cas9.
a, In order to introduce a single biotin moiety on Cas9, the solvent accessible, non-conserved amino-terminal methionine was mutated to a cysteine (M1C; red text) and the naturally occurring cysteine residues were mutated to serine (C80S and C574S; bold text). This enabled cysteine-specific labelling with EZ-link Maleimide-PEG2-biotin through an irreversible reaction between the reduced sulphydryl group of the cysteine and the maleimide group present on the biotin label. Mutations of dCas9 are also indicated in the domain schematic. b, Mass spectrometry analysis of the Cas9 biotin-labelling reaction confirmed that successful biotin labelling only occurs when the M1C mutation is present in the Cys-free background (C80S;C574S). The mass of the Maleimide-PEG2-biotin reagent is 525.6 Da. c, Streptavidin bead binding assay with biotinylated (biot.) or non-biotinylated (non-biot.) Cas9 and streptavidin agarose or streptavidin magnetic beads. Cas9 only remains specifically bound to the beads after biotin labelling. d, Cleavage assays were conducted as described in the Methods and resolved by denaturing PAGE. Reactions contained 100 nM Cas9 programmed with λ2 gRNA and ∼1 nM 5′-radiolabelled λ2 dsDNA target. e, Quantified cleavage data from triplicate experiments were fitted with single-exponential decays to calculate the apparent pseudo-first-order cleavage rate constants (average ± standard deviation). Both Cys-free and biotin-labelled Cas9(M1C) retain wild-type activity.
Extended Data Figure 6 RNA-guided Cas9 can utilize chemically modified PAMmers.
Nineteen-nucleotide PAMmer derivatives containing various chemical modifications on the 5′ and 3′ ends (capped) or interspersed throughout the strand still activate Cas9 for cleavage of ssRNA targets. These types of modification are often used to increase the in vivo half-life of short oligonucleotides by preventing exo- and endonuclease-mediated degradation. Cleavage assays were conducted as described in the Methods. PS, phosphorothioate bonds; LNA, locked nucleic acid.
Extended Data Figure 7 Cas9 programmed with GAPDH-specific gRNAs can pull down GAPDH mRNA in the absence of PAMmers.
a, Northern blot showing that, in some cases, Cas9–gRNA is able to pull down detectable amounts of GAPDH mRNA from total RNA without requiring a PAMmer. b, Northern blot showing that Cas9–gRNA G1 is also able to pull down quantitative amounts of GAPDH mRNA from HeLa cell lysate without requiring a PAMmer. s, standard; v1-5, increasingly 2′-OMe-modified PAMmers. See Fig. 4g for PAMmer sequences.
Extended Data Figure 8 Potential applications of RCas9 for untagged transcript analysis, detection and manipulation.
a, Catalytically active RCas9 could be used to target and cleave RNA, particularly those for which RNA-interference-mediated repression/degradation is not possible. b, Tethering the eukaryotic initiation factor eIF4G to a catalytically inactive dRCas9 targeted to the 5′ untranslated region of an mRNA could drive translation. c, dRCas9 tethered to beads could be used to specifically isolate RNA or native RNA–protein complexes of interest from cells for downstream analysis or assays including identification of bound-protein complexes, probing of RNA structure under native protein-bound conditions, and enrichment of rare transcripts for sequencing analysis. d, dRCas9 tethered to RNA deaminase or N6-mA methylase domains could direct site-specific A-to-I editing or methylation of RNA, respectively. e, dRCas9 fused to a U1 recruitment domain (arginine- and serine-rich (RS) domain) could be programmed to recognize a splicing enhancer site and thereby promote the inclusion of a targeted exon. f, dRCas9 tethered to a fluorescent protein such as GFP could be used to observe RNA localization and transport in living cells. Adapted from Mackay et al.18
Rights and permissions
About this article
Cite this article
O’Connell, M., Oakes, B., Sternberg, S. et al. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516, 263–266 (2014). https://doi.org/10.1038/nature13769
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13769
This article is cited by
-
Orthogonal inducible control of Cas13 circuits enables programmable RNA regulation in mammalian cells
Nature Communications (2024)
-
Programmable RNA detection with CRISPR-Cas12a
Nature Communications (2023)
-
Viral vectors and extracellular vesicles: innate delivery systems utilized in CRISPR/Cas-mediated cancer therapy
Cancer Gene Therapy (2023)
-
Simultaneous identification of viruses and viral variants with programmable DNA nanobait
Nature Nanotechnology (2023)
-
Variety Differentiation: Development of a CRISPR DETECTR Method for the Detection of Single Nucleotide Polymorphisms (SNPs) in Cacao (Theobroma cacao) and Almonds (Prunus dulcis)
Food Analytical Methods (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.