Abstract
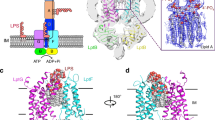
One of the fundamental properties of biological membranes is the asymmetric distribution of membrane lipids. In Gram-negative bacteria, the outer leaflet of the outer membrane is composed predominantly of lipopolysaccharides (LPS)1. The export of LPS requires seven essential lipopolysaccharide transport (Lpt) proteins to move LPS from the inner membrane, through the periplasm to the surface2. Of the seven Lpt proteins, the LptD–LptE complex is responsible for inserting LPS into the external leaflet of the outer membrane3,4. Here we report the crystal structure of the ∼110-kilodalton membrane protein complex LptD–LptE from Shigella flexneri at 2.4 Å resolution. The structure reveals an unprecedented two-protein plug-and-barrel architecture with LptE embedded into a 26-stranded β-barrel formed by LptD. Importantly, the secondary structures of the first two β-strands are distorted by two proline residues, weakening their interactions with neighbouring β-strands and creating a potential portal on the barrel wall that could allow lateral diffusion of LPS into the outer membrane. The crystal structure of the LptD–LptE complex opens the door to new antibiotic strategies targeting the bacterial outer membrane.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Muhlradt, P. F. & Golecki, J. R. Asymmetrical distribution and artifactual reorientation of lipopolysaccharide in the outer membrane bilayer of Salmonella typhimurium. Eur. J. Biochem. 51, 343–352 (1975)
Villa, R. et al. The Escherichia coli Lpt transenvelope protein complex for lipopolysaccharide export is assembled via conserved structurally homologous domains. J. Bacteriol. 195, 1100–1108 (2013)
Bos, M. P., Tefsen, B., Geurtsen, J. & Tommassen, J. Identification of an outer membrane protein required for the transport of lipopolysaccharide to the bacterial cell surface. Proc. Natl Acad. Sci. USA 101, 9417–9422 (2004)
Wu, T. et al. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl Acad. Sci. USA 103, 11754–11759 (2006)
Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 (2003)
Miller, S. I., Ernst, R. K. & Bader, M. W. LPS, TLR4 and infectious disease diversity. Nature Rev. Microbiol. 3, 36–46 (2005)
Ruiz, N., Kahne, D. & Silhavy, T. J. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nature Rev. Microbiol. 7, 677–683 (2009)
Ruiz, N., Gronenberg, L. S., Kahne, D. & Silhavy, T. J. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc. Natl Acad. Sci. USA 105, 5537–5542 (2008)
Sperandeo, P. et al. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J. Bacteriol. 190, 4460–4469 (2008)
Okuda, S., Freinkman, E. & Kahne, D. Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science 338, 1214–1217 (2012)
Ruiz, N., Chng, S. S., Hiniker, A., Kahne, D. & Silhavy, T. J. Nonconsecutive disulfide bond formation in an essential integral outer membrane protein. Proc. Natl Acad. Sci. USA 107, 12245–12250 (2010)
Chng, S. S. et al. Disulfide rearrangement triggered by translocon assembly controls lipopolysaccharide export. Science 337, 1665–1668 (2012)
Denoncin, K., Vertommen, D., Paek, E. & Collet, J. F. The protein-disulfide isomerase DsbC cooperates with SurA and DsbA in the assembly of the essential beta-barrel protein LptD. J. Biol. Chem. 285, 29425–29433 (2010)
Suits, M. D., Sperandeo, P., Deho, G., Polissi, A. & Jia, Z. Novel structure of the conserved gram-negative lipopolysaccharide transport protein A and mutagenesis analysis. J. Mol. Biol. 380, 476–488 (2008)
Tran, A. X., Dong, C. & Whitfield, C. Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J. Biol. Chem. 285, 33529–33539 (2010)
Srinivas, N. et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 327, 1010–1013 (2010)
Prilipov, A., Phale, P. S., Van Gelder, P., Rosenbusch, J. P. & Koebnik, R. Coupling site-directed mutagenesis with high-level expression: large scale production of mutant porins from E. coli. FEMS Microbiol. Lett. 163, 65–72 (1998)
Park, B. S. et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458, 1191–1195 (2009)
Chimalakonda, G. et al. Lipoprotein LptE is required for the assembly of LptD by the beta-barrel assembly machine in the outer membrane of Escherichia coli. Proc. Natl Acad. Sci. USA 108, 2492–2497 (2011)
Chng, S. S., Ruiz, N., Chimalakonda, G., Silhavy, T. J. & Kahne, D. Characterization of the two-protein complex in Escherichia coli responsible for lipopolysaccharide assembly at the outer membrane. Proc. Natl Acad. Sci. USA 107, 5363–5368 (2010)
Grabowicz, M., Yeh, J. & Silhavy, T. J. Dominant negative lptE mutation that supports a role for LptE as a plug in the LptD barrel. J. Bacteriol. 195, 1327–1334 (2013)
Freinkman, E., Chng, S. S. & Kahne, D. The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc. Natl Acad. Sci. USA 108, 2486–2491 (2011)
Braun, M. & Silhavy, T. J. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol. Microbiol. 45, 1289–1302 (2002)
van den Berg, B. Going forward laterally: transmembrane passage of hydrophobic molecules through protein channel walls. ChemBioChem 11, 1339–1343 (2010)
Khan, M. A. & Bishop, R. E. Molecular mechanism for lateral lipid diffusion between the outer membrane external leaflet and a beta-barrel hydrocarbon ruler. Biochemistry 48, 9745–9756 (2009)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Collaborative Computational Project, number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
DeLano, W. L. PyMOL molecular viewer: Updates and refinements. The 238th ACS National Meeting 238, (2009)
Acknowledgements
The authors thank Y. Shan for discussions, H. Wu, J. Deisenhofer, Y. Jiang, N. Huang, Z. Zhou, G. Li, Z. Liu and members of the Huang group for critically reading the manuscript. The diffraction data were collected at the Shanghai Synchrotron Radiation Facility (SSRF, China) and Beijing Synchrotron Radiation Facility of China (BSRF, China). This work was supported by grants from the Ministry of Science and Technology (2012CB917302 and 2013CB910603 to Y.H.), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB080203 to Y.H. and X.C.Z.) and the National Natural Science Foundation of China (31170698 to Y.H.).
Author information
Authors and Affiliations
Contributions
Y.H. supervised the project. S.Q. and Q.L. performed the experiments. S.Q., Y.Z. and Y.H. collected diffraction data. Y.H. built the model and refined the structure. Y.H., X.C.Z., S.Q. and Y.Z. contributed to manuscript preparation. Y.H. wrote the manuscript. All authors contributed to data analysis. Correspondence and material request should be address to Yihua Huang. The authors declare no competing financial interests.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Chemical structure and dimensions of LPS.
a, Chemical structure of LPS. LPS is composed of three modules: lipid A, a core oligosaccharide and a highly variable O-antigen constituted of repeating oligosaccharide units. The core is covalently linked to lipid A and can be further divided into inner and outer core. The inner core contains at least one residue of 3-deoxy-d-manno-oct-2 ulosonic acid (Kdo) linking to lipid A. b, Ra LPS is about 32 Å in height and 28 Å × 12 Å in the other two dimensions. The size of Ra LPS is based on the crystal structure of the TLR4/MD-2/Ra LPS complex (PDB ID 3FXI).
Extended Data Figure 2 Sequence alignment of LptD proteins from five representative Gram-negative bacterial species.
Secondary structures of LptD from S. flexneri (SfLptD) are labelled at the top of the sequence alignment. Strands that consist of the β-barrel of LptD are numbered. Residues of the two distorted β-strands (β1 and β2), L4 and L8 are boxed. Identical hydrophobic residues, positively-charged residues and negatively-charged residues are highlighted in yellow, blue and red, respectively. Three proline residues Pro 231, Pro 246 and Pro 261 in SfLptD are conserved in different species. SfLptD, StLptD, PaLptD, KpLptD and AbLptD represent LtpD sequences from S. flexneri, Salmonella enteric Typhimurium str. LT2, P. aeruginosa, Klebsiella pneumoniae and Acinetobacter baumannii, respectively.
Extended Data Figure 3 Two pairs of disulphide bonds are important for the stability and folding of LptD.
a, Non-reducing SDS–PAGE showing the purified LptD mutant proteins containing different Cys-to-Ser mutations. CCCC indicates that amino acids present at positions 31, 173, 724 and 725 in LptD are cysteine residues. LptD with mutation to Ser at different positions in the LptD sequence are co-expressed with LptE–His. b, Reducing SDS–PAGE showing LptD mutant proteins containing different Cys-to-Ser mutations. Samples were reduced by addition of 10 mM β-mercaptoethanol in the SDS–PAGE loading dye. All samples were boiled for 10 min at 95 °C. The degradation bands that appear below wild-type LptD were confirmed to be LptD fragments by mass spectrometry. All data shown are representative of at least three independent experiments.
Extended Data Figure 4 Structure-based sequence alignment of LptD_NT with LptA and LptC.
The three sequences show very low sequence identity, but have similar secondary structures and folds. The three LPS-binding residues in LptA and LptD_NT are boxed (‘×’ indicates residues that are invisible in the crystal structure).
Extended Data Figure 5 Electrostatic potential representation of the LptD–LptE complex.
a, Electrostatic surface of the LptD–LptE complex (side view). The hydrophobic girdle indicates the membrane-bound region, and this was verified by the presence of the expected aromatic belts. b, c, Periplasmic view of the interior surface properties of LptD–LptE complex (b) and that of LptD–LptE complex with LptD_NT domain removed (c). The interior surface of the complex is fairly hydrophilic with evenly distributed hydrophobic and hydrophilic residues lining inside.
Extended Data Figure 6 Structure superposition and sequence alignment of LptE orthologues.
a–c, Superposition of SfLptE with NmLptE (a), NeLptE (b) and SoLptE (c). d, Structure-based sequence alignment of SfLptE with NeLptE, NmLptE and SoLptE. SfLptD, NmLptE, NeLptE and SoLptE are coloured in magenta, orange, cyan and green, respectively. SfLptE, NeLptE, NmLptE and SoLptE represent LptE orthologues from S. flexneri, N. europaea, N. meningitides and S. oneidensis, respectively. Secondary structure elements are labelled on the top. The primary structures of these LptE orthologues have very low sequence identity, but their secondary structure distribution and fold are quite similar.
Extended Data Figure 7 Electron density maps showing detergent molecules in the LptD_NT groove and residues at the LPS exit portal.
a, Electron density maps (2Fo − Fc) showing the presence of two detergent molecules in the groove of LptD_NT. b, Stereo view of electron density map (2Fo − Fc) of residues at the potential LPS exit portal. The contour level for maps is 1.0σ.
Extended Data Figure 8 Residue E242 of LptD forms a polar-interaction network within strands β1 and β2.
Residue E242 of LptD forms a salt-bridge interaction with K234 and a few hydrogen bonds with residues of strands β1 and β2.
Extended Data Figure 9 Proposed model for LPS insertion into bacterial outer membrane.
LptD_NT relays LPS from LptA. Portion of LptD_NT of LptD likely resides in the outer membrane, and the LPS-conduit of the β-jellyroll is open to lipid phase (Fig. 1c). At the interface of LptD_NT domain and the β-barrel of LptD, the hydrophobic portion of LPS may directly enter into the lipid phase, whereas the hydrophilic moiety of LPS may enter into the barrel of LptD. The LPS molecules are inserted into the outer leaflet of the outer membrane upon dislocation of strands β1 and β2. LptE is a lipoprotein, and probably binds to the hydrophilic head groups of LPS in the process.
Rights and permissions
About this article
Cite this article
Qiao, S., Luo, Q., Zhao, Y. et al. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511, 108–111 (2014). https://doi.org/10.1038/nature13484
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13484
This article is cited by
-
Conformational Investigation of the Asymmetric Periplasmic Domains of E. coli LptB2FGC Using SDSL CW EPR Spectroscopy
Applied Magnetic Resonance (2023)
-
Modified Impedance Sensing System Determination of Virulence Characteristics of Pathogenic Bacteria Klebsiella Species
Indian Journal of Microbiology (2023)
-
Designing of multi-epitope peptide vaccine against Acinetobacter baumannii through combined immunoinformatics and protein interaction–based approaches
Immunologic Research (2023)
-
Physical properties of the bacterial outer membrane
Nature Reviews Microbiology (2022)
-
Antibacterial effects assessment on some livestock pathogens, thermal stability and proposing a probable reason for different levels of activity of thanatin
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.