Abstract
The Greenlandic population, a small and historically isolated founder population comprising about 57,000 inhabitants, has experienced a dramatic increase in type 2 diabetes (T2D) prevalence during the past 25 years1. Motivated by this, we performed association mapping of T2D-related quantitative traits in up to 2,575 Greenlandic individuals without known diabetes. Using array-based genotyping and exome sequencing, we discovered a nonsense p.Arg684Ter variant (in which arginine is replaced by a termination codon) in the gene TBC1D4 with an allele frequency of 17%. Here we show that homozygous carriers of this variant have markedly higher concentrations of plasma glucose (β = 3.8 mmol l−1, P = 2.5 × 10−35) and serum insulin (β = 165 pmol l−1, P = 1.5 × 10−20) 2 hours after an oral glucose load compared with individuals with other genotypes (both non-carriers and heterozygous carriers). Furthermore, homozygous carriers have marginally lower concentrations of fasting plasma glucose (β = −0.18 mmol l−1, P = 1.1 × 10−6) and fasting serum insulin (β = −8.3 pmol l−1, P = 0.0014), and their T2D risk is markedly increased (odds ratio (OR) = 10.3, P = 1.6 × 10−24). Heterozygous carriers have a moderately higher plasma glucose concentration 2 hours after an oral glucose load than non-carriers (β = 0.43 mmol l−1, P = 5.3 × 10−5). Analyses of skeletal muscle biopsies showed lower messenger RNA and protein levels of the long isoform of TBC1D4, and lower muscle protein levels of the glucose transporter GLUT4, with increasing number of p.Arg684Ter alleles. These findings are concomitant with a severely decreased insulin-stimulated glucose uptake in muscle, leading to postprandial hyperglycaemia, impaired glucose tolerance and T2D. The observed effect sizes are several times larger than any previous findings in large-scale genome-wide association studies of these traits2,3,4 and constitute further proof of the value of conducting genetic association studies outside the traditional setting of large homogeneous populations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Jørgensen, M. E., Bjerregaard, P. & Borch-Johnsen, K. Diabetes and impaired glucose tolerance among the Inuit population of Greenland. Diabetes Care 25, 1766–1771 (2002)
Scott, R. A. et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nature Genet. 44, 991–1005 (2012)
Morris, A. P. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nature Genet. 44, 981–990 (2012)
Manning, A. K. et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nature Genet. 44, 659–669 (2012)
Rafnar, T. et al. Mutations in BRIP1 confer high risk of ovarian cancer. Nature Genet. 43, 1104–1107 (2011)
Jørgensen, M. E., Borch-Johnsen, K., Stolk, R. & Bjerregaard, P. Fat distribution and glucose intolerance among Greenland Inuit. Diabetes Care 36, 2988–2994 (2013)
Voight, B. F. et al. The Metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 8, e1002793 (2012)
Marchini, J., Cardon, L. R., Phillips, M. S. & Donnelly, P. The effects of human population structure on large genetic association studies. Nature Genet. 36, 512–517 (2004)
Bjerregaard, P. et al. Inuit health in Greenland: a population survey of life style and disease in Greenland and among Inuit living in Denmark. Int. J. Circumpolar Health 62 (suppl. 1). 3–79 (2003)
Gutt, M. et al. Validation of the insulin sensitivity index (ISI0,120): comparison with other measures. Diabetes Res. Clin. Pract. 47, 177–184 (2000)
Cederberg, H. et al. Postchallenge glucose, A1C, and fasting glucose as predictors of type 2 diabetes and cardiovascular disease: a 10-year prospective cohort study. Diabetes Care 33, 2077–2083 (2010)
The 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010)
Lohmueller, K. E. et al. Whole-exome sequencing of 2,000 Danish individuals and the role of rare coding variants in type 2 diabetes. Am. J. Hum. Genet. 93, 1072–1086 (2013)
NHLBI. NHLBI GO Exome Sequencing Project (Exome Variant Server) http://evs.gs.washington.edu/EVS/
Sano, H. et al. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J. Biol. Chem. 278, 14599–14602 (2003)
Wang, H. Y. et al. AS160 deficiency causes whole-body insulin resistance via composite effects in multiple tissues. Biochem. J. 449, 479–489 (2013)
Baus, D. et al. Identification of a novel AS160 splice variant that regulates GLUT4 translocation and glucose-uptake in rat muscle cells. Cell. Signal. 20, 2237–2246 (2008)
Dash, S. et al. A truncation mutation in TBC1D4 in a family with acanthosis nigricans and postprandial hyperinsulinemia. Proc. Natl Acad. Sci. USA 106, 9350–9355 (2009)
Jørgensen, T. et al. A randomized non-pharmacological intervention study for prevention of ischaemic heart disease: baseline results Inter99. Eur. J. Cardiovasc. Prev. Rehabil. 10, 377–386 (2003)
Matthews, D. R. et al. Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28, 412–419 (1985)
World Health Organization Study Group. Definition, Diagnosis and Classification of Diabetes Mellitus and Its Complications. Part 1: Diagnosis and Classification of Diabetes Mellitus Technical Report Series WHO/NCD/NCS/99.2 (World Health Organization, 1999)
Zhou, X. & Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nature Genet. 44, 821–824 (2012)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010)
Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993 (2011)
Browning, B. L. & Browning, S. R. A unified approach to genotype imputation and haplotype-phase Inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84, 210–223 (2009)
Treebak, J. T. et al. Potential role of TBC1D4 in enhanced post-exercise insulin action in human skeletal muscle. Diabetologia 52, 891–900 (2009)
Albrechtsen, A. et al. Exome sequencing-driven discovery of coding polymorphisms associated with common metabolic phenotypes. Diabetologia 56, 298–310 (2013)
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009)
Moltke, I. & Albrechtsen, A. RelateAdmix: a software tool for estimating relatedness between admixed individuals. Bioinformatics 30, 1027–1028 (2014)
Acknowledgements
We thank I. Kleist, as well as all colleagues at the Physician’s Clinic, Nuuk, Greenland. We also thank J. F. Wojtaszewski for comments and X. Zhou for discussions about GEMMA. The Novo Nordisk Foundation Center for Basic Metabolic Research is an independent research centre at the University of Copenhagen and is partly funded by an unrestricted donation from the Novo Nordisk Foundation. This project was also funded by the Danish Council for Independent Research (Medical Sciences), the Steno Diabetes Center and the Villum Foundation. The IHIT study was supported by Karen Elise Jensen’s Foundation, NunaFonden, the Medical Research Council of Denmark, the Medical Research Council of Greenland and the Commission for Scientific Research in Greenland. None of the funding agencies had any role in the study design or in the collection or interpretation of the data.
Author information
Authors and Affiliations
Contributions
T.H. and A.A. conceived and headed the project. I.M., R.N. and A.A. designed the statistical set-up, and T.H., N.G., A.P.G. and O.P. designed the experimental set-up for the DNA extraction, genotyping and sequencing. A.L. provided the Danish samples. M.E.J., P.B. and K.B.-J. provided the Greenlandic samples, collected and defined the phenotypes and provided context for these samples. I.M. and A.A. performed the admixture, relatedness and linkage disequilibrium analyses. I.M. carried out the statistical part of the association analysis and N.G. carried out the medical part, with input from A.A., T.H., O.P. and R.N. The Chinese samples were analysed by X.W., G.S., X.J., J.A.-A. and J.W. N.G. analysed the Danish samples. X.W., G.S., X.J., J.A.-A. and J.W. performed the library constructions and sequencing. T.S.K. performed the mapping and genotyping for the sequencing data. M.F. and R.N. performed the selection analysis. N.T.K. collected muscle biopsies, and J.T.T., T.S.N., M.A.A. and J.R.Z. experimentally analysed and interpreted the TBC1D4 and GLUT4 expression data. N.G., I.M., A.A. and T.H. wrote most of the manuscript, with input from R.N., O.P., M.E.J. and P.B. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Linkage disequilibrium (LD) and relatedness in the IHIT cohort.
a, The mean LD at chromosome 13 as a function of distance estimated for a Danish population sample (Denmark) and for the IHIT cohort (Greenland). LD is measured in r2 values, and distance is measured as number of SNPs. b, A heat map of pairwise relatedness estimates based on the Metabochip SNP data from the individuals from the IHIT cohort and their estimated admixture proportions. k1 is an estimate of the fraction of sites where two individuals share one allele identity by descent (IBD), and k2 is an estimate of the fraction of sites where they share two alleles IBD. The estimates were achieved using the software tool RelateAdmix, which provides maximum likelihood estimates that take admixture into account by allowing alleles to be IBD only if the alleles are from the same population. The vast majority of the approximately 4 million pairwise estimates have a relatedness coefficient r of less than 1%, where r = 0.5k1 + k2.
Extended Data Figure 2 Association between fasting plasma glucose level, fasting serum insulin level and 2-h serum insulin level and the SNPs on the Metabochip for the IHIT cohort.
Analyses were done using an additive model and quantile-transformed phenotypes. Left, QQ plots with 95% confidence interval. The λ value is the genomic control inflation factor. Right, Manhattan plots showing the −log10[P] values. The red dashed horizontal line indicates the 0.05 significance threshold after Bonferroni correction for multiple testing.
Extended Data Figure 3 The region surrounding rs7330796.
a, Association results for all tested Metabochip SNPs in a 2-Mb region surrounding rs7330796 (shown as a dashed vertical line). Each SNP is represented by a coloured circle. The position of the circle along the x axis shows the genomic position of the SNP. The position of the circle along the left y axis shows the −log10[P] value of the SNP when testing for association with 2-h plasma glucose levels using an additive model. The colour of the circle indicates the r2 value between the SNP and rs7330796. The circle representing rs7330796 has a label to the left of it so that it can be identified. The solid blue curve (right y axis) illustrates the recombination rates from the Chinese HapMap (CHB) panel. Bottom, the protein-coding genes in the region are shown. b, Pairwise LD in the Greenlandic population for all SNPs with a MAF >0.05 in a 10-Mb region surrounding rs7330796. LD is measured in r2 and was estimated from the IHIT study sample.
Extended Data Figure 4 Mean 2-h plasma glucose levels in the IHIT cohort stratified by Inuit admixture proportion.
a, The three possible genotypes of rs7330796 (zero copies of the minor allele (WT), one copy (HE) and two copies (HO)). b, The three possible genotypes of p.Arg684Ter (zero copies of the p.Arg684Ter variant (WT), one copy (HE) and two copies (HO)). Error bars are s.e.m. and were estimated for each admixture proportion using a standard linear regression model.
Extended Data Figure 5 Selection analysis.
a, Haplotype structure of a 500-kilobase pair (kbp) region around the coding SNP rs61736969. For each SNP, the ancestral state is coloured red, while the derived allele is coloured yellow. Haplotypes are ordered according to the coding SNP rs61736969 (p.Arg684Ter), whose position is highlighted. Haplotypes above the black line carry the derived allele for rs61736969. b, The empirical distribution of the derived intra-allelic nucleotide diversity (DIND) values, computed on a 500-kbp region, for all SNPs with the same derived allele frequency as the coding SNP rs61736969. DIND is a measure of the ratio of the nucleotide diversity of haplotypes carrying the derived allele to the nucleotide diversity of haplotypes carrying the ancestral allele. The DIND value for the coding SNP rs61736969 is highlighted by a dashed line and lies in the top 5% of the distribution.
Extended Data Figure 6 Expression of TBC1D4 and GLUT4 in humans.
a, The mRNA expression levels of the long and short TBC1D4 isoforms in a range of different human tissues (measured in arbitrary units, a.u.). b, The mRNA expression levels of both TBC1D4 isoforms in human pancreatic islet cell preparations from five individuals (in a.u.). The five individuals are not Greenlandic and do not carry the p.Arg684Ter variant. The mean value for each of the isoforms is shown as a horizontal line. c, The mRNA expression levels of the short TBC1D4 isoform in skeletal muscle from nine Greenlanders: three with zero copies of the p.Arg684Ter variant (WT), three with one copy of the p.Arg684Ter variant (HE) and three with two copies of the p.Arg684Ter variant (HO). The arbitrary units (a.u.) are on the same scale as in Fig. 3d. The mean value for each genotype group is shown as a horizontal line. d, The protein abundance of GLUT4 in the same nine Greenlanders as in c (measured in a.u.). The mean value for each genotype group is shown as a horizontal line.
Supplementary information
Supplementary Notes
This file contains a brief description of the methods applied in selection analysis and the outcome of the analysis. (PDF 89 kb)
Rights and permissions
About this article
Cite this article
Moltke, I., Grarup, N., Jørgensen, M. et al. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature 512, 190–193 (2014). https://doi.org/10.1038/nature13425
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13425
This article is cited by
-
Wechselwirkung zwischen eingeschränkter Insulinsekretionskapazität und Insulinresistenz
Die Diabetologie (2024)
-
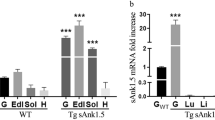
Deletion of Tbc1d4/As160 abrogates cardiac glucose uptake and increases myocardial damage after ischemia/reperfusion
Cardiovascular Diabetology (2023)
-
Similarity and diversity of genetic architecture for complex traits between East Asian and European populations
BMC Genomics (2023)
-
Dynamic measures of insulin action identify genetic determinants of dysglycemia
Nature Genetics (2023)
-
GWAS of lipids in Greenlanders finds association signals shared with Europeans and reveals an independent PCSK9 association signal
European Journal of Human Genetics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.