Abstract
Cancers have dysfunctional redox regulation resulting in reactive oxygen species production, damaging both DNA and free dNTPs. The MTH1 protein sanitizes oxidized dNTP pools to prevent incorporation of damaged bases during DNA replication. Although MTH1 is non-essential in normal cells, we show that cancer cells require MTH1 activity to avoid incorporation of oxidized dNTPs, resulting in DNA damage and cell death. We validate MTH1 as an anticancer target in vivo and describe small molecules TH287 and TH588 as first-in-class nudix hydrolase family inhibitors that potently and selectively engage and inhibit the MTH1 protein in cells. Protein co-crystal structures demonstrate that the inhibitors bind in the active site of MTH1. The inhibitors cause incorporation of oxidized dNTPs in cancer cells, leading to DNA damage, cytotoxicity and therapeutic responses in patient-derived mouse xenografts. This study exemplifies the non-oncogene addiction concept for anticancer treatment and validates MTH1 as being cancer phenotypic lethal.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
10 April 2014
Asterisks were missing in Fig. 5c and have now been added.
References
Druker, B. J. et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nature Med. 2, 561–566 (1996)
Bryant, H. E. et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose)polymerase. Nature 434, 913–917 (2005)
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005)
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013)
Zhang, Y. et al. Redox control of the survival of healthy and diseased cells. Antioxid. Redox Signal. 15, 2867–2908 (2011)
Luo, M., He, H., Kelley, M. R. & Georgiadis, M. M. Redox regulation of DNA repair: implications for human health and cancer therapeutic development. Antioxid. Redox Signal. 12, 1247–1269 (2010)
Topal, M. D. & Baker, M. S. DNA precursor pool: a significant target for N-methyl-N-nitrosourea in C3H/10T1/2 clone 8 cells. Proc. Natl Acad. Sci. USA 79, 2211–2215 (1982)
Sakumi, K. et al. Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J. Biol. Chem. 268, 23524–23530 (1993)
Oka, S. et al. Two distinct pathways of cell death triggered by oxidative damage to nuclear and mitochondrial DNAs. EMBO J. 27, 421–432 (2008)
Ichikawa, J. et al. Oxidation of mitochondrial deoxynucleotide pools by exposure to sodium nitroprusside induces cell death. DNA Repair (Amst.) 7, 418–430 (2008)
Russo, M. T. et al. The oxidized deoxynucleoside triphosphate pool is a significant contributor to genetic instability in mismatch repair-deficient cells. Mol. Cell. Biol. 24, 465–474 (2004)
Rai, P. et al. Enhanced elimination of oxidized guanine nucleotides inhibits oncogenic RAS-induced DNA damage and premature senescence. Oncogene 30, 1489–1496 (2011)
Tsuzuki, T., Egashira, A. & Kura, S. Analysis of MTH1 gene function in mice with targeted mutagenesis. Mutat. Res. 477, 71–78 (2001)
Schultz, L. B., Chehab, N. H., Malikzay, A. & Halazonetis, T. D. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 151, 1381–1390 (2000)
European Standards Committee on Oxidative DNA Damage Measurement of DNA oxidation in human cells by chromatographic and enzymic methods. Free Radic. Biol. Med. 34, 1089–1099 (2003)
Yoshimura, D. et al. An oxidized purine nucleoside triphosphatase, MTH1, suppresses cell death caused by oxidative stress. J. Biol. Chem. 278, 37965–37973 (2003)
Narita, M. et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113, 703–716 (2003)
Martinez Molina, D. et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013)
Svensson, L. M. et al. Crystal structure of human MTH1 and the 8-oxo-dGMP product complex. FEBS Lett. 585, 2617–2621 (2011)
Ushijima, Y. et al. A functional analysis of the DNA glycosylase activity of mouse MUTYH protein excising 2-hydroxyadenine opposite guanine in DNA. Nucleic Acids Res. 33, 672–682 (2005)
German, P. et al. Activation of cellular signaling by 8-oxoguanine DNA glycosylase-1-initiated DNA base excision repair. DNA Repair (Amst.) 12, 856–863 (2013)
Hahn, W. C. et al. Creation of human tumour cells with defined genetic elements. Nature 400, 464–468 (1999)
McLennan, A. G. Substrate ambiguity among the nudix hydrolases: biologically significant, evolutionary remnant, or both? Cell. Mol. Life Sci. 70, 373–385 (2013)
Maki, H. & Sekiguchi, M. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 355, 273–275 (1992)
Fujikawa, K. et al. The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein. J. Biol. Chem. 274, 18201–18205 (1999)
Speina, E. et al. Contribution of hMTH1 to the maintenance of 8-oxoguanine levels in lung DNA of non-small-cell lung cancer patients. J. Natl Cancer Inst. 97, 384–395 (2005)
Guo, Z., Kozlov, S., Lavin, M. F., Person, M. D. & Paull, T. T. ATM activation by oxidative stress. Science 330, 517–521 (2010)
Huber, K. V. M. et al. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature http://dx.doi.org/10.1038/nature13194 (this issue)
De Luca, G. et al. Prolonged lifespan with enhanced exploratory behavior in mice overexpressing the oxidized nucleoside triphosphatase hMTH1. Aging Cell 12, 695–705 (2013)
De Luca, G. et al. A role for oxidized DNA precursors in Huntington's disease-like striatal neurodegeneration. PLoS Genet. 4, e1000266 (2008)
Sakumi, K. et al. Ogg1 knockout-associated lung tumorigenesis and its suppression by Mth1 gene disruption. Cancer Res. 63, 902–905 (2003)
Mohindra, A. et al. Defects in homologous recombination repair in mismatch-repair-deficient tumour cell lines. Hum. Mol. Genet. 11, 2189–2200 (2002)
Rai, P. et al. Continuous elimination of oxidized nucleotides is necessary to prevent rapid onset of cellular senescence. Proc. Natl Acad. Sci. USA 106, 169–174 (2009)
Herold, M. J., van den Brandt, J., Seibler, J. & Reichardt, H. M. Inducible and reversible gene silencing by stable integration of an shRNA-encoding lentivirus in transgenic rats. Proc. Natl Acad. Sci. USA 105, 18507–18512 (2008)
O’Brien, J., Wilson, I., Orton, T. & Pognan, F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur. J. Biochem. 267, 5421–5426 (2000)
Struthers, L., Patel, R., Clark, J. & Thomas, S. Direct detection of 8-oxodeoxyguanosine and 8-oxoguanine by avidin and its analogues. Anal. Biochem. 255, 20–31 (1998)
Dalle-Donne, I., Rossi, R., Giustarini, D., Milzani, A. & Colombo, R. Protein carbonyl groups as biomarkers of oxidative stress. Clin. Chim. Acta 329, 23–38 (2003)
Baykov, A. A., Evtushenko, O. A. & Avaeva, S. M. A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal. Biochem. 171, 266–270 (1988)
Britsun, V. M. & Maiboroda, E. I. Cyclocondensations of 3-(R2-amino)-3-methylthio-1-R1-propenones with 2-aminoazoles. Ukr. Khim. Zh. 74, 126–128 (2008)
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Waters, N. J., Jones, R., Williams, G. & Sohal, B. Validation of a rapid equilibrium dialysis approach for the measurement of plasma protein binding. J. Pharm. Sci. 97, 4586–4595 (2008)
Houston, J. B. Utility of in vitro drug metabolism data in predicting in vivo metabolic clearance. Biochem. Pharmacol. 47, 1469–1479 (1994)
Baranczewski, P. et al. Introduction to in vitro estimation of metabolic stability and drug interactions of new chemical entities in drug discovery and development. Pharmacol. Rep. 58, 453–472 (2006)
Hubatsch, I., Ragnarsson, E. G. & Artursson, P. Determination of drug permeability and prediction of drug absorption in Caco-2 monolayers. Nature Protocols 2, 2111–2119 (2007)
Acknowledgements
We thank scientists at BESSY, ESRF, Max-Lab and the Swiss Light Source for structural biology data collection support, PSF for protein purification, GE Healthcare for instrument access, HTSRC (Rockefeller University) for iTC200 calorimeter access, S. Nordstrand for animal support, and L. Ny, U. Stierner and J. Mattsson for discussions. The Helleday Laboratory is primarily funded by the Torsten and Ragnar Söderberg Foundation (T.H.). This project is primarily supported by The Knut and Alice Wallenberg Foundation. Further support was received from the Swedish Research Council (T.H., A.J.J., P.A., P.S., J.A.N.), the European Research Council (T.H.), Swedish Cancer Society (T.H., J.A.N.), the Swedish Children’s Cancer Foundation (T.H.), AFA insurance (T.H.), the Swedish Pain Relief Foundation (T.H.), The Cancer Society in Stockholm (T.H.), the Wenner-Gren Foundations (P.S.), the Swedish Foundation for Strategic Research (P.S.), the Dutch Cancer Society (B.E.), EMBO LTF (R.B.), Region Västra Götaland (J.A.N.), BioCARE (J.A.N.), the Swiss National Science Foundation (F.Z.G.) and the Nicholson Exchange Program (T.L.). Chemical Biology Consortium Sweden (CBCS) is primarily funded by the Swedish Research Council. CBCS acknowledge Swedish Orphan Biovitrum for their donation of a small-molecule infrastructure including a compound collection.
Author information
Authors and Affiliations
Contributions
T.H. devised concept and supervised the project. H.G., S.E., N.S., C.E.S., L.B., K.Sa., F.Jo., A.Hö., B.E., T.D., M.A., A.Ha., C.L., U.W.B. and T.H. designed, performed and analysed cell biology experiments. A-S.J., T.L., F.Je., O.L., K.St., T.K., M.Hä., U.M., B.L., L.J, A.J.J., E.W., C.K., I.A., S.A.J., U.W.B. and T.H. designed, performed and analysed biochemical and high-throughput experiments. L.M.S., R.P.-A.B., R.G. and P.S. designed, performed and analysed structural biology experiments. T.K., S.A.J., M.D., M.-C.J.-C., L.J., L.G.J.H., M.He, K.S.A.V., O.A.W., A.J.J., M.S. and T.H. designed, performed and analysed medicinal chemistry experiments. A.-L.G., J.C.P., E.J.H. and M.D. performed computational chemistry analysis and/or support. C.G., B.O.E., A.S., K.Sa., P.B., R.S., F.Z.G., I.G., P.A., T.P., S.V., J.A.N. and U.W.B. designed, performed and analysed ADME, pharmacology and in vivo experiments. R.O. performed surgery, and clinical follow-up, H.G., U.W.B. and T.H. wrote the paper. All authors discussed results and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
A patent has been filed with data generated in this manuscript where T.H., M.S., T.K., S.A.J., M.D., M-C.JC. are listed as inventors.
Extended data figures and tables
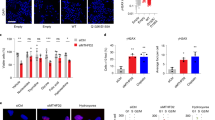
Extended Data Figure 1 Transient siRNA depletion of MTH1 reduces cell survival and causes DNA damage.
a–d, U2OS cells were transfected with siRNA for 3 days, after which (a) cell viability was determined after additional 3 days in culture; (b) cells were trypsinized, counted and re-seeded for clonogenic outgrowth; (c) proteins extracted and analysed with western blot (data shown as average ± s.d. from three independent experiments for a–c); or (d) grown for additional 3 days and immunoblotted with 53BP1 and RPA-32 antibodies and analysed with immunofluorescence. DNA was counterstained with ToPro-3 iodide. e, f, Quantification of 53BP1 (e) (data shown as average ± s.e.m. from three independent experiments, *P < 0.05; **P < 0.01, one-way ANOVA) and RPA foci (f) (percentage of cells >5 foci per cell) after MTH1 siRNA transfection. Data shown as average ± s.e.m. from two experiments. *P < 0.05, one-way ANOVA. g, Transient siRNA depletion of MTH1 reduces cell survival to various extents in cancer cell lines. Indicated cells were transfected with MTH1 siRNA #3 (M) and non-targeting control (N) for 3 days. The cells were trypsinized, counted and re-seeded for clonogenic outgrowth. Values represent relative survival of MTH1 siRNA-transfected cells compared to non-targeting control. Data shown as average ± s.d. from two independent experiments. h, The cells were collected, proteins extracted and analysed with western blot. Figure shows one representative western blot from two independent experiments. MTH1 knockdown by siRNA induces cell death in most cancer cells, but not in normal cells (VH10, MRC-9, BJ-hTERT).
Extended Data Figure 2 Rescue expression of RNAi-resistant wild-type MTH1 and catalytically dead MTH1(E56A) mutant and purification of MTH1 protein and activity measurements.
a, U2OS cells stably overexpressing RNAi-resistant wild-type (WT) MTH1 and catalytically dead mutant (E56A) were transfected for 72 h, and western blot of MTH1 protein levels in non-targeting siRNA (N), MTH1 siRNA (M), MTH1 wild type (WT) or MTH1 E56A is shown. Figure shows one representative western blot from three independent experiments. b, The MTH1 wild type (WT) or MTH1 E56A were expressed in E. coli and purified to >98% purity. c, Relative activity of MTH1 wild type and E56A catalysed hydrolysis of dGTP, demonstrating that the E56A is a catalytically dead mutant of MTH1. Data shown as a representative graph, average ± s.d. from two independent experiments, each performed in triplicate. d–f, SDS–PAGE gels showing (d) the purity of the MTH1 W117Y and D119A mutant proteins; (e) the purity of expressed and purified OGG1; and (f) the purity of expressed and purified MUTYH in fusion with MBP (maltose binding protein). g, Activity of MTH1 mutants D119A and W117Y relative to wild type with 20 μM 8-oxodGTP and 2-OHdATP. Data shown as average ± s.d. from two independent experiments, each performed in triplicates. h, U2OS cells stably overexpressing RNAi-resistant wild-type MTH1 and MTH1 mutants (D119A and W117Y) were transfected for 72 h and western blot of MTH1 protein levels in non-targeting siRNA (N) and MTH1 siRNA (M) is shown. Figure shows one representative western blot from two independent experiments. i, Clonogenic survival of U2OS cells transiently transfected with MTH1 siRNA (M) or NT siRNA (N). Cells were transfected for 72 h and reseeded for clonogenic outgrowth. Values represent average ± s.d. from two independent experiments. j, Determination of carbonylated proteins in primary VH10 and U2OS cancer cells. Values represent average ± s.e.m. from four independent experiments. Statistically significant P = 0.033 in Student’s t-test. k, Immunofluorescent staining of phospho-p53 (S15) and phospho-ATM (S1981) in U2OS cells transiently transfected with NT siRNA or MTH1 siRNA. l, ATM phosphorylation, p53 phosphorylation and p21 induction is induced by MTH1 siRNA depletion (M) in U2OS cells, but not in NT siRNA control (N). Incubation with ATM inhibitor KU55933 (10 μM) prevents ATM and p53 phosphorylation after MTH1 siRNA depletion as well as induction of p21. m, The second independent experiment using MTH1 shRNA as in Fig. 2l. Doxycycline was added to the drinking water (indicated by the arrow) (n = 7 per group, data shown as average ± s.e.m). n, Kaplan–Meyer survival plot of mice carrying SW480 tumours with the indicated construct and addition of doxycycline to the drinking water. At a volume size of 1,000 mm3, animals were euthanized. All animals in the control groups, MTH1 shRNA without doxycycline and NT shRNA with doxycycline, had reached the upper limit of tumour size at 45 and 32 days, respectively, whereas the MTH1 shRNA knockdown group was all still alive at day 62.
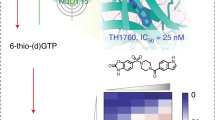
Extended Data Figure 3 Screening for MTH1 inhibitors and MTH1 protein binding.
a, Purity of MTH1 and MutT preparations used. b, High-throughput screening identified hits containing 2-aminopyrimidine motif (scaffold 1). By introducing an aminomethyl substituent a marked increase in potency was observed (TH086). The dichlorophenyl derivative TH287 showed further improved potency, however, with a high intrinsic clearance in mouse liver microsomes. The cyclopropyl compound TH588 displayed increased metabolic stability while retaining potency against MTH1. (IC50 values, determined using the sensitive version of the malachite-green-based enzymatic assay, are displayed as average ± s.d., n = 3 independent experiments.) c, Representative dose-response curves of scaffold 1, TH086, TH287 and TH588. d, e, 2-OHdATP (d) or 8-oxodGTP (e) was used as substrates showing that the inhibition of MTH1 by TH588 and TH287 is substrate independent. (Graphs in c, d and e show representative curves, average ± s.d. from two replicates; IC50 values were determined using the PPiLight Inorganic Pyrophosphate Assay kit (Lonza) and are presented as average ± s.d. from two independent experiments.) f, To confirm binding of optimized hits to MTH1 protein, ITC experiments were performed. Left-hand graphs show raw data (black) from two independent titrations of TH086 (129 μM) into a solution containing MTH1 protein (30 μM). The right-hand graph shows the integrated heats from the titrations of TH086 into a solution containing MTH1 as a function of their molar ratio. The solid lines represent the best-fit binding isotherms to the data (one set of sites model within the Origin software), K = 6.1 ± 1.5 × 107 M−1, ΔH = −59 ± 0.5 kJ per mole and n = 0.71 (blue) and K = 9.2 ± 2.2 × 107 M−1, ΔH = −58 ± 0.4 kJ per mole and n = 0.70 (red), respectively. g, h, Biacore surface plasmon resonance technique was used to determine the affinity of (g) TH287 (average ± s.d.: Kd = 1.7 ± 0.6 nM, n = 3) and (h) TH588 (average ± s.d.; Kd = 15.5 ± 2.9 nM, n = 3) to the MTH1 protein. Representative sensorgram for each inhibitor is shown (n = 3 independent experiments). Data points confounded by the buffer change during the start and end of injection are excluded from the analysis and the graphic representation. i, j, Electron density for (i) TH287 and (j) TH588. 2Fo − Fc maps contoured at 1.5σ.
Extended Data Figure 4 TH588 reduces tumour growth in mouse xenografts.
a, Pharmacokinetic profiles of TH287 (10 mg kg−1, s.c.). b, TH588 pharmacokinetic profile (30 mg kg−1, s.c.) (average ± s.e.m. (n = 3 per data point) in a, b). (See also Extended Data Table 2.) c, Chemical structure and IC50 value of TH287’s and TH588’s major metabolite TH586. d, e, TH588 (30 mg kg−1 s.c., once daily (q.d.)) significantly reduced tumour growth in SW480 xenograft mice after 35 days treatment (average ± s.e.m., n = 8 per group, two-way ANOVA with repeated measures; *P < 0.05, **P < 0.01) (d), with no effect on body weight (e). f, Tiled images of SW480 xenograft with start at the capsule (left) of the tumours. Green stain shows Ki67, red stain shows CD31, blue stain shows DNA signal. Scale bar, 1 mm. i:1, untreated xenograft; i:3, xenograft treated with TH588. i:2 and i:4 show close-ups of the marked area in i:1 and i:3. Scale bar, 100 μm. g, h, Vascularization (CD31), measured as the area of CD31 staining normalized against area of DNA (g), and proliferation (Ki67), measured as the mean signal of Ki67 in areas with DNA (h), in untreated and TH588-treated xenograft in relation to the distance from the capsule of the tumours (average ± s.e.m. untreated, n = 7 for TH588 treated, n = 8 for untreated g, h). i, j, TH588 (30 mg kg−1 s.c., q.d) significantly reduced tumour growth in MCF-7 xenograft (i) (average ± s.e.m., n = 10 per group, two-way ANOVA with repeated measures; *P < 0.05) with no effect on body weight (j). k, Clinical parameters measured in blood from SW480 xenograft animals in d, e. The mean values of white blood cells (WBC), red blood cells (RBC), neutrophils, lymphocytes, monocytes, mean corpuscular volume (MCV), mean cell haemoglobin (MCH), mean cell haemoglobin concentration (MCHC) from the different groups are presented in the table. The results did not show any significant differences in the haematology parameters or the liver/heart/kidney parameters between control and treated groups.
Extended Data Figure 5 MTH1 inhibitors TH588 and TH287 induce oxidative lesions in the DNA and kill cancer cells.
a–c, U2OS cells (a, b) or VH10 cells (c) were treated with 10 μM MTH1 inhibitor for 24 h before being incubated with MUTYH or with buffer alone (control) and run in alkaline comet assay. As a positive control, cells were treated with 100 μM H2O2 for 5 min on ice. Tail moment is calculated as per cent DNA in the tail multiplied by the tail length. Values represent average ± s.e.m. from three experiments; *P < 0.05; **P < 0.01; ***P < 0.001, one-way ANOVA. d, e, Viability determined by resazurin in U2OS cells expressing wild-type MTH1, MTH1(W117Y) or MTH1(D119A) exposed 72 h to increasing concentrations of MTH1 inhibitors (d) TH287 or (e) TH588. Data are shown as average ± s.e.m. from three independent experiments. f, g, Viability of U2OS cells stably carrying doxycycline-inducible p53 shRNA constructs after 72 h exposure to different concentrations of MTH1 inhibitors (f) TH287 or (g) TH588. Data shown as average ± s.e.m. from two independent experiments. h, i, Viability of HCT116 and HCT116 p53−/− after 72 h exposure to different concentrations of MTH1 inhibitors (h) TH287 or (i) TH588. Values represent percentage of cells in relation to mock-treated controls displayed as average ± s.d. from three independent experiments. j, Cell viability after MTH1 inhibitor treatment determined by resazurin assay in primary or immortalized cells or in cancer cell lines. All data are average ± s.e.m. of at least three independent experiments. ND, not determined. k, l, Viability determined in melanoma cancer cell lines after exposure for 72 h to increasing concentrations of MTH1 inhibitors (k) TH287 or (l) TH588. Values represent percentage of cells in relation to mock-treated controls displayed as mean ± s.d. from three independent experiments, each done in duplicate. m, Effect of TH287 on clonogenic cell survival in primary or immortalized cells (open) or in cancer cell lines (filled). n, No toxicity was observed in clonogenic survival of TH650 in various cell lines. o–q, Clonogenic survival of BJ cells after treatment with (o) TH588, (p) TH287, and (q) TH650. TH588 and TH287 both significantly reduce cell survival primarily in hTERT immortalized BJ cells transfected with SV40 large T (Sv40T) or SV40 large T and Ras (RasV12), whereas no toxicity was observed with the less potent MTH1 inhibitor TH650. For m–q: cells were seeded in 6-well plates (200 cells per well for primary/immortalized cells, except BJ-hTERT) or in 10-cm dishes (500 cells per dish for all other cells) in triplicates and the day after treated with 0–10 μM compound for 7–10 days after which the number of colonies were counted. Values represent percentage of colonies in relation to DMSO-treated controls displayed as average ± s.d. from three independent experiments.
Extended Data Figure 6 MTH1 inhibitors induce DNA damage foci in U2OS but not in VH10 cells.
a–l, Cells were seeded on coverslips (a–c), 96-well plates (d–h) or 6-well plates (i–l) and were the following day treated with TH287, TH588 and TH650. DMSO concentration was kept constant. a–h, Samples were fixed with formaldehyde after an additional 72 h and immunostained with (a, d) 53BP1, (b–f) RPA and (c, g, h) DNA-PKcs pS2056 antibodies followed by secondary Alexa-conjugated antibodies. The DNA was counterstained with (a–c) To-Pro3 or (d–h) DAPI. Images were acquired and the number of foci was quantified by automatic image analysis. Scale bar, 10 μm. Data shown as average ± s.e.m. from three (d, g, h) or four (e, f) independent experiments. i–l, Cells were washed with PBS and detached by scraping in lysis buffer. i, j, Samples were analysed on western blot with ATM phospho-S1981, p53 phospho-S15, p21 and α-tubulin antibodies. k, l, FACS analysis of apoptosis in U2OS cells treated with TH287 and TH588 for 72 h. k, Quantification of the percentage of cleaved caspase 3 positive cells. l, Propidium iodine staining and quantification of percentage of cells in sub-G1 phase. Data shown as average ± s.e.m. from two independent experiments performed in duplicate. Blots shown as a representative blot from two experiments.
Extended Data Figure 7 Expression level of MTH1, MTH2, MUTYH or OGG1 in cell lines used in this study.
a, Heat-map diagram of MTH1, MTH2, MUTYH and OGG1 gene expression determined by qRT–PCR in the same cell lines. The expression levels are normalized to VH10 cells. ΔΔCq value was calculated using Bio-Rad CFX Manager 3. The normalized levels below 1 have lighter green colour than VH10. b, MTH1 protein levels determined by western blot analysis of cell lines used in this study. c, Quantification of MTH1 protein levels depicted as MTH1/actin ratio and normalized to VH10 cells, in primary or immortalized cells (green) or in cancer cells (red) reveals no obvious correlation between protein levels and sensitivity to MTH1 inhibitors or siRNA response. Values represent average ± s.d. from two experiments, except for cell line 1BR2 for which values represent one experiment. d–g, The bar chart display of panel a. Values represent average ± s.e.m. from two experiments performed in triplicate. h, MTH1 (NUDT1) expression in normal and cancer tissues (from http://www.genesapiens.org).
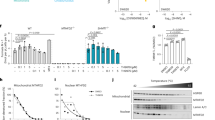
Extended Data Figure 8 Expression analysis, MTH1 inhibitor selectivity and MutT overexpression.
a, Expression level of MTH1 after doxycycline-induced depletion in SW480 cells with MTH1 shRNA construct. b–d, Quantification of MTH2 (NUDT15), MUTYH or OGG1 expression levels using qRT–PCR in (b) non-targeting (NT) shRNA, or doxycycline treatment for 48 h (c) or 96 h (d). Values represent average ± s.e.m. from three experiments performed in triplicate. ΔΔCq value was calculated using Bio-Rad CFX Manager 3. The expression levels were normalized to VH10 cells. e, Overexpression of OGG1 or MUTYH in U2OS cells determined by qRT–PCR expression. Values represent average ± s.e.m. from two independent experiments, each performed in triplicate. ΔΔCq value was calculated using Bio-Rad CFX Manager 3. The expressions were normalized to the CMV-empty control. *P < 0.05, Student’s t-test by Bio-Rad CFX manager 3. f, Survival in U2OS overexpressing either MUTYH or OGG1 after 72 h treatment with TH588. Values represent average ± s.d. from three independent experiments, each performed in triplicate. g, Selectivity of TH588 against a panel of diverse GPCRs, ion channels, transporters and enzymes, including ALK kinase. TH588 (10 μM) was tested at CEREP high-throughput screening panel to obtain information about possible cross-reactivity. Notably, TH588 did not show any significant inhibitory effect on hERG. Additional functional studies need to be performed before a full evaluation of selectivity of the compound can be made. h, Purity of enzyme preparations used in the selectivity screen. 4 μg of the enzyme preparations used were analysed using SDS–PAGE followed by Coomassie staining. i, Inhibition analysis of TH287 showing >1,000 fold selectivity for MTH1. Data shown as average ± s.d., n = 3–6 independent experiments performed in duplicate. j, Inhibition analysis of TH650 showing >100 fold selectivity for MTH1. Data shown as average ± s.d., n = 3–6 independent experiments performed in duplicate. k, MutT overexpression in U2OS cells. Western blot of U2OS cells stably transfected with MutT-MLS-myc (MutT-m), MutT-myc + MutT-MLS-myc (MutT-mn) or no expression (control) immunoblotted with Myc and tubulin antibodies. l, Immunofluorescence image of U2OS cells transiently transfected with MutT-mn and immunostained with Myc antibody. m, Immunofluorescence image of U2OS cells transiently transfected with MutT-m and immunostained with Myc and ATP synthase antibodies followed by secondary Alexa-conjugated antibody. DNA was counterstained with To-Pro3. Scale bar, 20 μm.
Supplementary information
Supplementary Methods
This file contains details of the synthesis of specific MTH1 inhibitors. (PDF 309 kb)
Rights and permissions
About this article
Cite this article
Gad, H., Koolmeister, T., Jemth, AS. et al. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature 508, 215–221 (2014). https://doi.org/10.1038/nature13181
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13181
This article is cited by
-
Therapeutic targeting nudix hydrolase 1 creates a MYC-driven metabolic vulnerability
Nature Communications (2024)
-
NUDT22 promotes cancer growth through pyrimidine salvage
Oncogene (2023)
-
MTH1 protects platelet mitochondria from oxidative damage and regulates platelet function and thrombosis
Nature Communications (2023)
-
Mammalian Nudt15 hydrolytic and binding activity on methylated guanosine mononucleotides
European Biophysics Journal (2023)
-
Tips for efficiently maintaining pET expression plasmids
Current Genetics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.