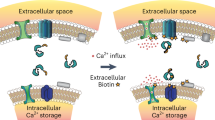
Abstract
Primary cilia are solitary, non-motile extensions of the centriole found on nearly all nucleated eukaryotic cells between cell divisions. Only ∼200–300 nm in diameter and a few micrometres long, they are separated from the cytoplasm by the ciliary neck and basal body. Often called sensory cilia, they are thought to receive chemical and mechanical stimuli and initiate specific cellular signal transduction pathways. When activated by a ligand, hedgehog pathway proteins, such as GLI2 and smoothened (SMO), translocate from the cell into the cilium1,2. Mutations in primary ciliary proteins are associated with severe developmental defects3. The ionic conditions, permeability of the primary cilia membrane, and effectiveness of the diffusion barriers between the cilia and cell body are unknown. Here we show that cilia are a unique calcium compartment regulated by a heteromeric TRP channel, PKD1L1–PKD2L1, in mice and humans. In contrast to the hypothesis that polycystin (PKD) channels initiate changes in ciliary calcium that are conducted into the cytoplasm4, we show that changes in ciliary calcium concentration occur without substantially altering global cytoplasmic calcium. PKD1L1–PKD2L1 acts as a ciliary calcium channel controlling ciliary calcium concentration and thereby modifying SMO-activated GLI2 translocation and GLI1 expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Corbit, K. C. et al. Vertebrate Smoothened functions at the primary cilium. Nature 437, 1018–1021 (2005)
Huangfu, D. & Anderson, K. V. Cilia and Hedgehog responsiveness in the mouse. Proc. Natl Acad. Sci. USA 102, 11325–11330 (2005)
Goetz, S. C. & Anderson, K. V. The primary cilium: a signalling centre during vertebrate development. Nature Rev. Genet. 11, 331–344 (2010)
Praetorius, H. A. & Spring, K. R. Bending the MDCK cell primary cilium increases intracellular calcium. J. Membr. Biol. 184, 71–79 (2001)
DeCaen, P. G., Delling, M., Vien, T. N. & Clapham, D. E. Direct recording and molecular identification of the calcium channel of primary cilia. Nature http://dx.doi.org/10.1038/nature12832 (this issue)
Hama, H. et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nature Neurosci. 14, 1481–1488 (2011)
Tian, L. et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nature Methods 6, 875–881 (2009)
Yasuda, R. et al. Imaging calcium concentration dynamics in small neuronal compartments. Sci. STKE 2004, pl5 (2004)
Soler-Llavina, G. J. & Sabatini, B. L. Synapse-specific plasticity and compartmentalized signaling in cerebellar stellate cells. Nature Neurosci. 9, 798–806 (2006)
Clapham, D. E. Calcium signaling. Cell 131, 1047–1058 (2007)
Vogel, P. et al. Situs inversus in Dpcd/Poll−/−, Nme7−/−, and Pkd1l1−/− mice. Vet. Pathol. 47, 120–131 (2010)
Field, S. et al. Pkd1l1 establishes left-right asymmetry and physically interacts with Pkd2. Development 138, 1131–1142 (2011)
Horio, N. et al. Sour taste responses in mice lacking PKD channels. PLoS ONE 6, e20007 (2011)
Martin, V. & Shaw-Smith, C. Review of genetic factors in intestinal malrotation. Pediatr. Surg. Int. 26, 769–781 (2010)
Ramalho-Santos, M., Melton, D. A. & McMahon, A. P. Hedgehog signals regulate multiple aspects of gastrointestinal development. Development 127, 2763–2772 (2000)
Humke, E. W., Dorn, K. V., Milenkovic, L., Scott, M. P. & Rohatgi, R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 24, 670–682 (2010)
Keady, B. T. et al. IFT25 links the signal-dependent movement of Hedgehog components to intraflagellar transport. Dev. Cell 22, 940–951 (2012)
Kim, J., Kato, M. & Beachy, P. A. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc. Natl Acad. Sci. USA 106, 21666–21671 (2009)
Bhogaraju, S., Taschner, M., Morawetz, M., Basquin, C. & Lorentzen, E. Crystal structure of the intraflagellar transport complex 25/27. EMBO J. 30, 1907–1918 (2011)
Zhao, Y. et al. An expanded palette of genetically encoded Ca2+ indicators. Science 333, 1888–1891 (2011)
Miki, K. & Clapham, D. E. Rheotaxis guides mammalian sperm. Curr. Biol. 23, 443–452 (2013)
Gibbs, D. & Williams, D. S. Isolation and culture of primary mouse retinal pigmented epithelial cells. Adv. Exp. Med. Biol. 533, 347–352 (2003)
Preibisch, S., Saalfeld, S. & Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465 (2009)
Kim, J. et al. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature 464, 1048–1051 (2010)
Tukachinsky, H., Lopez, L. V. & Salic, A. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J. Cell Biol. 191, 415–428 (2010)
Acknowledgements
We thank the Mouse Gene Manipulation Facility of the Boston Children’s Hospital Intellectual and Developmental Disabilities Research Center (IDDRC; NIHP30-HD 18655). We thank the Image and Data Analysis Core (IDAC) at Harvard Medical School for help with three-dimensional reconstruction. We also thank A. Salic for anti-SMO and anti-GLI2 antibodies. P.G.D. was supported by NIH T32-HL007572. We thank M. Desai for graphical assistance and A. von Gise and H. Tukachinsky (Harvard Medical School) and the members of the Clapham laboratory for advice and discussion.
Author information
Authors and Affiliations
Contributions
All authors designed or conducted experiments and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 ARL13B–EGFP identifies primary and motile cilia in transgenic mouse tissue.
a–l, Frozen tissue sections were prepared from P0 intestine (a–c); E15 nasal cavity (d–f); 6-week-old hippocampus (g–i); P0 retina from Arl13b-EGFPtg mice (j–l). First column, Arl13b-EGFPtg (green) fluorescence; second column (red) labels villin in b, ACIII in e, h, or HMB-45 (human melanoma black antibody, retinal pigmented epithelial cells) in k. a–c, Intestine: EGFP fluorescence in a does not overlap with the anti-villin staining in b as shown in the merged image in c, indicating that ARL13B–EGFP is absent from microvilli (arrows). Scale bar: 100 μm. d–f, Nasal cavity: (d) several ARL13B–EGFP-positive cilia (arrows) that face the lumen of the turbinate co-localize with ACIII (e) as shown in the merged image (f). Scale bar: 100 μm. Inset shows magnification of nasal cavity surface. Scale bar of inset in f, 5 μm. g–i, CA1 region of hippocampus: (g) prominent cilia (arrows) overlap with ACIII staining (h) as shown in the merged image (i). Scale bar: 50 μm. Inset shows magnification of hippocampal cilia. Inset scale bar: 5 µm. Merged red and green channels were offset for clarity (inset). j–l, Retinal pigmented epithelia (RPE): (j) short cilia (arrow) are visible at the intersection between RPE (labelled with HMB-45 antibody) and (k) developing photoreceptor cells, as shown in the merged image (l). Scale bar: 10 μm. Inset shows magnification of RPE–photoreceptor interface. Inset scale bar: 2 μm.
Extended Data Figure 2 ARL13B–EGFP labels a primary cilium in cultured MEF and RPE cells.
a–l, The first column shows ARL13B–EGFPtg (green) fluorescence; second column labels ACIII (b, e) or acetylated tubulin (h, k). Third column: merged red and green channels were offset for clarity. Primary MEFs of Arl13b-EGFPtg (a–c) and wild-type (d–f) mice isolated from E14.5 embryos. ARL13B–EGFP co-localizes with ciliary ACIII in c. MEFs isolated from wild-type mice show no fluorescence in the cilium (488 nm excitation). g–i, Primary RPE cells isolated from P12 Arl13b-EGFPtg mice. Cells were fixed and stained with antibody to acetylated tubulin. ARL13B–EGFP (g) exclusively localized to the primary cilium identified by antibody to acetylated tubulin (h, i). j–l, Stable cell line (hRPE1) expressing SMO–EGFP. After 2 days of serum starvation, SMO–EGFP labelled the primary cilium of hRPE1 cells (j) as indicated by acetylated tubulin labelling (k, l). Scale bars: c, f, i, l, 5 μm. m, n, To determine whether ARL13B–EGFP expression adversely affected ciliogenesis, ciliary length and per cent of cells with cilia were quantified from wild-type and ARL13B–EGFP-expressing MEFs stained by anti-acetylated tubulin. m, Ciliary length was similar in wild-type and ARL13B–EGFP-expressing MEFs (2.6 ± 0.5 μm versus 2.9 ± 0.8 μm, respectively, n = 200). n, The number of cells with cilia was also comparable between wild-type and ARL13B–EGFP-expressing MEFs (60.2% ± 5.1% versus 65.5% ± 7.3%; n = 120).
Extended Data Figure 3 Ciliary [Ca2+] changes in stably transfected RPE cell cilia.
a–c, Live hRPE1 cells stably expressing SMO–GCaMP3 were treated with 5 μM ionomycin. Fluorescence increases were measured in the cilium and cytoplasm. Image in c is DIC. d, Changes in fluorescence (ΔF/F) of the calcium sensor, GCaMP3, are plotted for both cytoplasm and cilium. e–h, hRPE1 cells expressing SMO–mCherry–GCaMP3 were stained with acetylated tubulin. GCaMP3 (e) and mCherry (f) fluorescence overlaps with acetylated tubulin staining (g, h). Scale bar: 5 μm; merged channels were offset for clarity. i, The tip of a cilium was ruptured with an intense 1-s laser pulse (405 nm, hRPE1 cell expressing SMO–mCherry–GCaMP3). Circle indicates area of rupture. Numbered arrowheads indicate positions where changes in fluorescence were measured. Asterisk indicates mCherry fluorescence outside of cilium, indicating that some SMO–mCherry–GCaMP3 is retained in the ER. j, Quantification of changes in fluorescence at the positions marked in i. Rupture of the ciliary membrane rapidly increases [Ca2+]cilia at the tip and travels along the cilium at 4.6 ± 0.6 µm s−1 (n = 16).
Extended Data Figure 4 [Ca2+]cilia calibration.
a, Images of hRPE1 cells stably expressing SMO–mCherry–GCaMP3 were acquired after permeabilization with 15 μM digitonin in varying extracellular [Ca2+]. b, Averages of several ratios (n = 12–16; ±s.d.) per concentration are plotted against free [Ca2+], yielding the calibration curve for SMO–mCherry–GCaMP3: Kd = 625 nM.
Extended Data Figure 5 Overexpressed and endogenous PKD2L1 localizes to the primary cilium.
a–c, Rabbit anti-PKD2L1 (Thermo Scientific) recognizes overexpressed PKD2L1. HEK cells were transfected with hPKD2L1-IRES mCherry construct and stained with PKD2L1 antibody. PKD2L1 staining (a) is specific to cells that also express mCherry (b). c, Overlay. d–f, Overexpressed hPKD2L1 localizes to the primary cilium in mIMCD3 cells. mIMCD3 cells were transfected with HA-tagged hPKD2L1 and stained with an anti-HA antibody (d) and anti-acetylated tubulin antibody (e). HA immunoreactivity is visible both in the cytoplasm and in the cilium. f, Overlay. g, h, PKD2L1 antibody labels the primary cilium of mIMCD3 cells. g, Confluent mIMCD3 cells were stained with anti-PKD2L1 antibody used in a; and h, acetylated tubulin antibody to label cilia. PKD2L1 immunoreactivity is visible in the primary cilium. i, Overlay. j–o, Primary MEFs of Arl13b-EGFPtg (j–l) and Arl13b-EGFPtg × Pkd2l1−/− mice (m–o) isolated from E14.5 embryos were stained with anti-PKD2L1 antibody used in a. In Arl13b-EGFPtg MEFs, PKD2L1 immunoreactivity (j) co-localizes with ARL13B–EGFP signal (k) labelling the primary cilium. l, Overlay. m–o, PKD2L1 staining is absent in cilia of Arl13b-EGFPtg × Pkd2L1−/− mice. Scale bars: 10 μm.
Extended Data Figure 6 [Ca2+]cilia increases 24 h after SAG stimulation.
a, MEFs expressing ARL13B–mCherry–GECO1.2 were stained with acetylated tubulin. GECO1.2 and mCherry fluorescence overlaps with acetylated tubulin staining. Scale bar: 5 μm; merged channels were offset for clarity. b, Ratio maps of MEFs isolated from ARL13B–mCherry–GECO1.2 mice stimulated with 0.05% DMSO (left) or 400 nM SAG (right) for 24 h. Scale bar: 5 μm. c, Quantification of ciliary GECO1.2/mCherry ratios obtained for MEFs with and without SAG stimulation. Ratio increases from 0.4 ± 0.05 to 0.8 ± 0.2 after SAG stimulation (*P < 0.05; n = 20–30 cilia). d, Example ciliary current measured from MEFs treated with 500 nM SAG (SMO agonist) or with DMSO vehicle (0.05%) in culture for 24–36 h in control conditions and after activation with 10 μM calmidazolium (CMZ). e, Scatter and whisker (±s.d.) plots from cilia show total outward (+100 mV) and inward (−100 mV) current measured for both treatment groups. Averages are indicated by the thick horizontal lines and individual cilium current magnitudes are represented as circles. P values resulting from Student’s t-test comparing treatment groups are indicated (*< 0.05; n = 11 cilia).
Supplementary information
Oviductal motile cilia from an 8-week-old Arl13B-EGFPtg mouse
The oviduct was isolated after hormone treatment (Methods), opened longitudinally, and mounted on a coverslip. Motile cilia were imaged at a frame rate of 65 ms. Scale bar, 5 µm. Cilia sweep oviductal debris from left to right. (MOV 3935 kb)
3D projection of cilia expressing Arl13B-EGFP
Rotating 3D projection of the middle digit of an E14.5 Arl13B-EGFPtg mouse paw showing all cilia in this developing tissue. Scale bar, 200 µm. (MOV 94206 kb)
Smo-GCaMP3 reports [Ca2+] changes in cilium and cytoplasm
Addition of 5 µM Ionomycin to Smo-GCaMP3 expressing hRPE1 cell increases fluorescence in the cilium and cytoplasm. Scale bar, 5 µm. Recorded at 250 ms per frame. (MOV 932 kb)
: Laser rupture of a cilium expressing Smo-mCherry-GCaMP3
The tip of a cilium was ruptured by a 1-2 s pulse from a 405 nm laser. Calcium influx is visible at the right tip of the cilium. The resulting increase in [Ca2+] diffuses along the cilium. From a Smo-mCherry-GCaMP3-expressing hRPE1 cell recorded at 65 ms per frame. Scale bar, 5 µm. (MOV 1783 kb)
Ciliary [Ca2+] can increase independently of the cytoplasm
The cytoplasm of a Smo-mCherry-GCaMP3-expressing hRPE1 cell was loaded with Fluo-4 Ca2+ indicator (Fluo-4-AM, 10 µM) and fluorescence measured in the cilium and cytoplasm. The tip of a cilium was then ruptured by a 1-2 s pulse from a 405 nm laser. 2 mM [Ca2+] entering the cilium overwhelms the ciliary buffers, saturates the ciliary Smo-mCherry-GCaMP3 reporter, and after an ~ 40 s delay, barely increases cytoplasmic [Ca2+] (t= 59 s). Ionomycin added to the bath at 1 min 20 s results in a very large increase in the cytoplasm. Recorded at 1.1 s per frame. Scale bar, 5 µm. (MOV 413 kb)
Cytoplasmic [Ca2+] rises propagate into the cilium
The cytoplasm of a Smo-mCherry-GCaMP3-expressing hRPE1 cell was loaded with o-Nitrophenyl EGTA AM (NP-EGTA AM, 10 µM) and fluorescence measured in the cilium and cytoplasm. Both GCaMP3 (left) and mCherry (right) fluorescence are shown. Calcium was uncaged in the cytoplasm by a 1 s pulse from a 405 nm laser. A calcium wave travels inside the cytoplasm and enters the cilium. Scale bar, 5 µm. (MOV 5836 kb)
Rights and permissions
About this article
Cite this article
Delling, M., DeCaen, P., Doerner, J. et al. Primary cilia are specialized calcium signalling organelles. Nature 504, 311–314 (2013). https://doi.org/10.1038/nature12833
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12833
This article is cited by
-
The multifaceted links between hearing loss and chronic kidney disease
Nature Reviews Nephrology (2024)
-
Keeping pace: the primary cilium as the conducting baton of the islet
Diabetologia (2024)
-
1-Indanone retards cyst development in ADPKD mouse model by stabilizing tubulin and down-regulating anterograde transport of cilia
Acta Pharmacologica Sinica (2023)
-
Glucose absorption drives cystogenesis in a human organoid-on-chip model of polycystic kidney disease
Nature Communications (2022)
-
A change of heart: new roles for cilia in cardiac development and disease
Nature Reviews Cardiology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.