Abstract
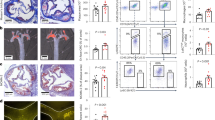
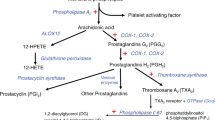
Myocardial infarction, a leading cause of death in the Western world1, usually occurs when the fibrous cap overlying an atherosclerotic plaque in a coronary artery ruptures. The resulting exposure of blood to the atherosclerotic material then triggers thrombus formation, which occludes the artery2. The importance of genetic predisposition to coronary artery disease and myocardial infarction is best documented by the predictive value of a positive family history3. Next-generation sequencing in families with several affected individuals has revolutionized mutation identification4. Here we report the segregation of two private, heterozygous mutations in two functionally related genes, GUCY1A3 (p.Leu163Phefs*24) and CCT7 (p.Ser525Leu), in an extended myocardial infarction family. GUCY1A3 encodes the α1 subunit of soluble guanylyl cyclase (α1-sGC)5, and CCT7 encodes CCTη, a member of the tailless complex polypeptide 1 ring complex6, which, among other functions, stabilizes soluble guanylyl cyclase. After stimulation with nitric oxide, soluble guanylyl cyclase generates cGMP, which induces vasodilation and inhibits platelet activation7. We demonstrate in vitro that mutations in both GUCY1A3 and CCT7 severely reduce α1-sGC as well as β1-sGC protein content, and impair soluble guanylyl cyclase activity. Moreover, platelets from digenic mutation carriers contained less soluble guanylyl cyclase protein and consequently displayed reduced nitric-oxide-induced cGMP formation. Mice deficient in α1-sGC protein displayed accelerated thrombus formation in the microcirculation after local trauma. Starting with a severely affected family, we have identified a link between impaired soluble-guanylyl-cyclase-dependent nitric oxide signalling and myocardial infarction risk, possibly through accelerated thrombus formation. Reversing this defect may provide a new therapeutic target for reducing the risk of myocardial infarction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Data deposits
Variant data is available in ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) with accession numbers SCV000083870 for NM_001130683.2:c.488dup and SCV000083871 for NM_001166284.1:c.1313C>T.
References
World Health Organization. The top 10 causes of death. Fact sheet no. 310; http://www.who.int/mediacentre/factsheets/fs310/en/index.html (2011)
Finn, A. V., Nakano, M., Narula, J., Kolodgie, F. D. & Virmani, R. Concept of vulnerable/unstable plaque. Arterioscler. Thromb. Vasc. Biol. 30, 1282–1292 (2010)
Lloyd-Jones, D. M. et al. Parental cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults: a prospective study of parents and offspring. J. Am. Med. Assoc. 291, 2204–2211 (2004)
Bamshad, M. J. et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nature Rev. Genet. 12, 745–755 (2011)
Mergia, E., Friebe, A., Dangel, O., Russwurm, M. & Koesling, D. Spare guanylyl cyclase NO receptors ensure high NO sensitivity in the vascular system. J. Clin. Invest. 116, 1731–1737 (2006)
Leitner, A. et al. The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure 20, 814–825 (2012)
Zabel, U., Weeger, M., La, M. & Schmidt, H. H. Human soluble guanylate cyclase: functional expression and revised isoenzyme family. Biochem. J. 335, 51–57 (1998)
The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010)
Moro, M. A. et al. cGMP mediates the vascular and platelet actions of nitric oxide: confirmation using an inhibitor of the soluble guanylyl cyclase. Proc. Natl Acad. Sci. USA 93, 1480–1485 (1996)
Deloukas, P. et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nature Genet. 45, 25–33 (2013)
Lu, X. et al. Genome-wide association study in Han Chinese identifies four new susceptibility loci for coronary artery disease. Nature Genet. 44, 890–894 (2012)
Stasch, J. P., Pacher, P. & Evgenov, O. V. Soluble guanylate cyclase as an emerging therapeutic target in cardiopulmonary disease. Circulation 123, 2263–2273 (2011)
Broeckel, U. et al. A comprehensive linkage analysis for myocardial infarction and its related risk factors. Nature Genet. 30, 210–214 (2002)
Fischer, M. et al. Distinct heritable patterns of angiographic coronary artery disease in families with myocardial infarction. Circulation 111, 855–862 (2005)
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nature Methods 7, 248–249 (2010)
Risch, N. Linkage strategies for genetically complex traits. III. The effect of marker polymorphism on analysis of affected relative pairs. Am. J. Hum. Genet. 46, 242–253 (1990)
Dekker, C. et al. The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins. EMBO J. 30, 3078–3090 (2011)
Krieger, E. et al. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins 77, 114–122 (2009)
Krieger, E., Darden, T., Nabuurs, S. B., Finkelstein, A. & Vriend, G. Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins 57, 678–683 (2004)
Krieger, E., Nielsen, J. E., Spronk, C. A. & Vriend, G. Fast empirical pK a prediction by Ewald summation. J. Mol. Graph. Model. 25, 481–486 (2006)
Liu, Y., Ruoho, A. E., Rao, V. D. & Hurley, J. H. Catalytic mechanism of the adenylyl and guanylyl cyclases: modeling and mutational analysis. Proc. Natl Acad. Sci. USA 94, 13414–13419 (1997)
Wölfle, S. E., Schmidt, V. J., Hoyer, J., Kohler, R. & de Wit, C. Prominent role of KCa3.1 in endothelium-derived hyperpolarizing factor-type dilations and conducted responses in the microcirculation in vivo . Cardiovasc. Res. 82, 476–483 (2009)
Schunkert, H. et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nature Genet. 43, 333–338 (2011)
Preuss, U. W., Koller, G., Zill, P., Bondy, B. & Soyka, M. Alcoholism-related phenotypes and genetic variants of the CB1 receptor. Eur. Arch. Psychiatry Clin. Neurosci. 253, 275–280 (2003)
Ritsch, A. et al. Cholesteryl ester transfer protein and mortality in patients undergoing coronary angiography: the Ludwigshafen Risk and Cardiovascular Health study. Circulation 121, 366–374 (2010)
Haenle, M. M. et al. Overweight, physical activity, tobacco and alcohol consumption in a cross-sectional random sample of German adults. BMC Public Health 6, 233 (2006)
Schunkert, H. et al. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation 117, 1675–1684 (2008)
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature Genet. 39, 906–913 (2007)
Acknowledgements
We thank all the family members who participated in this research. Without the continuous support of these patients over more than 15 years, the present work would not have been possible. We would like to thank S. Wrobel, S. Stark, A. Liebers, K. Franke, J. Stegmann-Frehse, M. Behrensen, M. Schmid, J. Eckhold, D. Wöllner, U. Krabbe and J. Simon for technical assistance. Furthermore, we would like to thank M. Becker, N. Buchholz, I. Demuth, R. Eckardt, H. Heekeren, U. Lindenberger, M. Lövdén, L. Müller, W. Nietfeld, G. Pawelec, F. Schmiedeck, T. Siedler and G. G. Wagner for their contributions to BASE-II. We also would like to thank S. Herterich and S. Gambaryan for advice, and B. Mayer, U. Hubauer, K.-H. Ameln and A. Großhennig for help with GerMIFS. We thank WTCCC+ and the WTCCC-CAD2 investigators for access to their data. The study is supported by the Deutsche Forschungsgemeinschaft and the German Federal Ministry of Education and Research (BMBF) in the context of the German National Genome Research Network (NGFN-2 (01GS0417) and NGFN-plus (01GS0832)), the FP6 and FP7 EU-funded integrated projects Cardiogenics (LSHM-CT-2006-037593), ENGAGE (201413), and GEUVADIS (261123), the binational BMBF/ANR funded project CARDomics (01KU0908A), the local focus programs ‘Kardiovaskuläre Genomforschung’ and ‘Medizinische Genetik’ of the Universität zu Lübeck, and the University Hospital of Regensburg, Germany. The German Federal Ministry for Education and Research provided funding for BASE-II (BMBF; grant no. 16SV5538). Support by NSFC grant 30730057 from the Chinese Government (to J.O.) is gratefully acknowledged. N.J.S. holds a Chair funded by the British Heart Foundation, and is supported by the Leicester NIHR Biomedical Research Unit in Cardiovascular Disease. U.W. is supported by the BMBF (01EO1003). M.M.N. is a member of the DFG-funded Excellence Cluster ImmunoSensation.
Author information
Authors and Affiliations
Consortia
Contributions
J.E., C.H., F.J.K., T.M., N.J.S., H.S. and K.S. designed the study. Z.A., D.B., P.B., C.d.W., S.E., U.B.E., E.G., F.J.K., D.K., A.M., E.M., W.R., P.M.R., T.M.S. and M.E.Z. conducted the experiments. I.B., M.F., J.N., J.O., K.S., T.M.S., S.T. and C.W. analysed the data. A.J.B., L.B., P.S.B., C.B., A.S.H., P.H., S.K., S.-C.L., W.M., R.M., T.M., M.M.N., M.R., N.J.S., S.S., E.S.-T. and U.W. provided material, data and analysis tools. J.E., C.d.W., C.H., F.J.K., N.J.S. and H.S. wrote the paper. C.H. and H.S. contributed equally. See Supplementary Information for Members of CARDIoGRAM.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A list of authors and their affiliations appears in the Supplementary Information.
Supplementary information
Supplementary Information
This file contains Supplementary Materials and Methods, Supplementary Tables 1-7, Supplementary Figures 1-8 and funding and affiliation details for CARDIoGRAM. (PDF 3117 kb)
Thrombus formation in arterioles in vivo
An arteriole in cremaster-microcirculation with a diameter of about 40 µm. Blood flow is from bottom to top, the animal has received high molecular weight FITC-dextran intravenously which is photoexcited by illuminating the preparation at 450-490nm using mercury lamp at time point 01:07:00. This is visible as a light spot in arteriole, which reflects the excitation of the dye. Thrombus formation starts to be visible downstream of the excitated area in the upper right corner 42 s later. Thereafter, flow decelerates and comes to complete stop 88 s after starting the photoexcitation, which is turned-off at this point. (AVI 7116 kb)
Rights and permissions
About this article
Cite this article
Erdmann, J., Stark, K., Esslinger, U. et al. Dysfunctional nitric oxide signalling increases risk of myocardial infarction. Nature 504, 432–436 (2013). https://doi.org/10.1038/nature12722
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12722
This article is cited by
-
Biallelic variants in NOS3 and GUCY1A3, the two major genes of the nitric oxide pathway, cause moyamoya cerebral angiopathy
Human Genomics (2023)
-
Loss of soluble guanylyl cyclase in platelets contributes to atherosclerotic plaque formation and vascular inflammation
Nature Cardiovascular Research (2022)
-
Toxoplasmosis: Targeting neurotransmitter systems in psychiatric disorders
Metabolic Brain Disease (2022)
-
Nitrate-functionalized patch confers cardioprotection and improves heart repair after myocardial infarction via local nitric oxide delivery
Nature Communications (2021)
-
Genetische Diagnostik in der Kardiologie - wann ist es sinnvoll?
CardioVasc (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.