Abstract
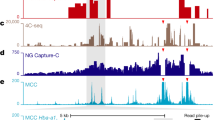
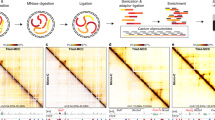
A large number of cis-regulatory sequences have been annotated in the human genome1,2, but defining their target genes remains a challenge3. One strategy is to identify the long-range looping interactions at these elements with the use of chromosome conformation capture (3C)-based techniques4. However, previous studies lack either the resolution or coverage to permit a whole-genome, unbiased view of chromatin interactions. Here we report a comprehensive chromatin interaction map generated in human fibroblasts using a genome-wide 3C analysis method (Hi-C)5. We determined over one million long-range chromatin interactions at 5–10-kb resolution, and uncovered general principles of chromatin organization at different types of genomic features. We also characterized the dynamics of promoter–enhancer contacts after TNF-α signalling in these cells. Unexpectedly, we found that TNF-α-responsive enhancers are already in contact with their target promoters before signalling. Such pre-existing chromatin looping, which also exists in other cell types with different extracellular signalling, is a strong predictor of gene induction. Our observations suggest that the three-dimensional chromatin landscape, once established in a particular cell type, is relatively stable and could influence the selection or activation of target genes by a ubiquitous transcription activator in a cell-specific manner.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Accessions
Gene Expression Omnibus
Data deposits
All sequencing data described in this study have been deposited to GEO under the accession number GSE43070. Some sequencing data used in this study were previously published and accession numbers can be found in Supplementary Methods. All chromatin interactions called in IMR90 cells can be found in Supplementary Data.
References
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012)
Maurano, M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012)
Smallwood, A. & Ren, B. Genome organization and long-range regulation of gene expression by enhancers. Curr. Opin. Cell Biol. 25, 387–394 (2013)
Dekker, J., Marti-Renom, M. A. & Mirny, L. A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nature Rev. Genet. 14, 390–403 (2013)
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009)
Imakaev, M. et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nature Methods 9, 999–1003 (2012)
de Wit, E. & de Laat, W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 26, 11–24 (2012)
Mercer, T. R. et al. DNase I-hypersensitive exons colocalize with promoters and distal regulatory elements. Nature Genet. 45, 852–859 (2013)
Noordermeer, D. et al. The dynamic architecture of Hox gene clusters. Science 334, 222–225 (2011)
Lettice, L. A. et al. Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc. Natl Acad. Sci. USA 99, 7548–7553 (2002)
Zhang, Y. et al. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 148, 908–921 (2012)
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012)
Sexton, T. et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012)
Hawkins, R. D. et al. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell 6, 479–491 (2010)
Sanyal, A., Lajoie, B. R., Jain, G. & Dekker, J. The long-range interaction landscape of gene promoters. Nature 489, 109–113 (2012)
Creyghton, M. P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA 107, 21931–21936 (2010)
Hawkins, R. D. et al. Dynamic chromatin states in human ES cells reveal potential regulatory sequences and genes involved in pluripotency. Cell Res. 21, 1393–1409 (2011)
Rada-Iglesias, A. et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 (2011)
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010)
Phillips-Cremins, J. E. et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153, 1281–1295 (2013)
Francis, N. J., Kingston, R. E. & Woodcock, C. L. Chromatin compaction by a polycomb group protein complex. Science 306, 1574–1577 (2004)
Thurman, R. E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012)
Ong, C. T. & Corces, V. G. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nature Rev. Genet. 12, 283–293 (2011)
Schoenfelder, S., Clay, I. & Fraser, P. The transcriptional interactome: gene expression in 3D. Curr. Opin. Genet. Dev. 20, 127–133 (2010)
Melo, C. A. et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol. Cell 49, 524–535 (2013)
Tan, P. Y. et al. Integration of regulatory networks by NKX3–1 promotes androgen-dependent prostate cancer survival. Mol. Cell. Biol. 32, 399–414 (2012)
Jin, F., Li, Y., Ren, B. & Natarajan, R. P. U. 1 and C/EBP(alpha) synergistically program distinct response to NF-κB activation through establishing monocyte specific enhancers. Proc. Natl Acad. Sci. USA 108, 5290–5295 (2011)
John, S. et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nature Genet. 43, 264–268 (2011)
Mullen, A. C. et al. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell 147, 565–576 (2011)
Jin, F., Li, Y., Ren, B. & Natarajan, R. Enhancers: multi-dimensional signal integrators. Transcription 2, 226–230 (2011)
Acknowledgements
We thank C. K. Glass for sharing the GRO-seq protocol, and S. Kuan and L. Edsall for assistance with high-throughput DNA sequencing and the initial processing. This work is supported by funds from the Ludwig Institute for Cancer Research, the California Institute of Regenerative Medicine (RN2-00905) and US National Institutes of Health (P50 GM085764-03 and U01 ES017166).
Author information
Authors and Affiliations
Contributions
Y.L., F.J. and B.R. designed the studies. Y.L. conducted most of the experiments; F.J. carried out the data analysis; J.R.D., Z.Y., A.Y.L., C.Y., A.D.S. and C.E. contributed to the experiments; S.S. contributed to the data analysis; F.J., Y.L. and B.R. prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Tables 1-6, Supplementary References and Supplementary Figures 1-24. (PDF 3678 kb)
Supplementary Data 1
This file contains data sheets of mapped cis-elements in IMR90 cells used in this study, including genomic locations of active TSS’s, inactive TSS’s, active enhancers, poised enhancers, CTCF peaks, H3K27me3 peaks and p65 peaks. (XLSX 5022 kb)
Supplementary Data 2
This file contains data sheets of chromatin interactions looping to the active promoters in IMR90 cells. The first sheet lists the locations of 11,313 anchors covering all active promoters in IMR90 cells. The second sheet lists the locations of all target peaks interacting with these anchors. (XLSX 3837 kb)
Supplementary Data 3
This zipped file contains a text file listing the locations of all 518,032 anchors covering every HindIII fragment in the human genome. Chromatin interactions were called for every one of these anchors. (ZIP 4820 kb)
Supplementary Data 4
This zipped file contains a text file listing the location of all 1,116,312 chromatin interactions identified in this study. (ZIP 19585 kb)
Rights and permissions
About this article
Cite this article
Jin, F., Li, Y., Dixon, J. et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013). https://doi.org/10.1038/nature12644
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12644
This article is cited by
-
Enhancer–promoter interactions become more instructive in the transition from cell-fate specification to tissue differentiation
Nature Genetics (2024)
-
Increased enhancer–promoter interactions during developmental enhancer activation in mammals
Nature Genetics (2024)
-
A spatial genome aligner for resolving chromatin architectures from multiplexed DNA FISH
Nature Biotechnology (2023)
-
Enhancer-instructed epigenetic landscape and chromatin compartmentalization dictate a primary antibody repertoire protective against specific bacterial pathogens
Nature Immunology (2023)
-
Superenhancers as master gene regulators and novel therapeutic targets in brain tumors
Experimental & Molecular Medicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.