Abstract
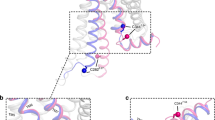
G-protein-coupled receptors (GPCRs) are integral membrane proteins that have an essential role in human physiology, yet the molecular processes through which they bind to their endogenous agonists and activate effector proteins remain poorly understood. So far, it has not been possible to capture an active-state GPCR bound to its native neurotransmitter. Crystal structures of agonist-bound GPCRs have relied on the use of either exceptionally high-affinity agonists1,2 or receptor stabilization by mutagenesis3,4,5. Many natural agonists such as adrenaline, which activates the β2-adrenoceptor (β2AR), bind with relatively low affinity, and they are often chemically unstable. Using directed evolution, we engineered a high-affinity camelid antibody fragment that stabilizes the active state of the β2AR, and used this to obtain crystal structures of the activated receptor bound to multiple ligands. Here we present structures of the active-state human β2AR bound to three chemically distinct agonists: the ultrahigh-affinity agonist BI167107, the high-affinity catecholamine agonist hydroxybenzyl isoproterenol, and the low-affinity endogenous agonist adrenaline. The crystal structures reveal a highly conserved overall ligand recognition and activation mode despite diverse ligand chemical structures and affinities that range from 100 nM to ∼80 pM. Overall, the adrenaline-bound receptor structure is similar to the others, but it has substantial rearrangements in extracellular loop three and the extracellular tip of transmembrane helix 6. These structures also reveal a water-mediated hydrogen bond between two conserved tyrosines, which appears to stabilize the active state of the β2AR and related GPCRs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rosenbaum, D. M. et al. Structure and function of an irreversible agonist–β2 adrenoceptor complex. Nature 469, 236–240 (2011)
Xu, F. et al. Structure of an agonist-bound human A2A adenosine receptor. Science 332, 322–327 (2011)
White, J. F. et al. Structure of the agonist-bound neurotensin receptor. Nature 490, 508–513 (2012)
Lebon, G. et al. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature 474, 521–525 (2011)
Warne, T. et al. The structural basis for agonist and partial agonist action on a β1-adrenergic receptor. Nature 469, 241–244 (2011)
Venkatakrishnan, A. J. et al. Molecular signatures of G-protein-coupled receptors. Nature 494, 185–194 (2013)
Nygaard, R. et al. The dynamic process of β2-adrenergic receptor activation. Cell 152, 532–542 (2013)
Rasmussen, S. G. et al. Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature 477, 549–555 (2011)
Rasmussen, S. G. et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature 469, 175–180 (2011)
Caffrey, M. & Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nature Protocols 4, 706–731 (2009)
Deupi, X. et al. Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc. Natl Acad. Sci. USA 109, 119–124 (2012)
Schneider, E. H., Schnell, D., Strasser, A., Dove, S. & Seifert, R. Impact of the DRY motif and the missing “ionic lock” on constitutive activity and G-protein coupling of the human histamine H4 receptor. J. Pharmacol. Exp. Ther. 333, 382–392 (2010)
Elgeti, M. et al. Conserved Tyr2235.58 plays different roles in the activation and G-protein interaction of rhodopsin. J. Am. Chem. Soc. 133, 7159–7165 (2011)
Goncalves, J. A. et al. Highly conserved tyrosine stabilizes the active state of rhodopsin. Proc. Natl Acad. Sci. USA 107, 19861–19866 (2010)
Serrano-Vega, M. J., Magnani, F., Shibata, Y. & Tate, C. G. Conformational thermostabilization of the β1-adrenergic receptor in a detergent-resistant form. Proc. Natl Acad. Sci. USA 105, 877–882 (2008)
Strader, C. D., Candelore, M. R., Hill, W. S., Sigal, I. S. & Dixon, R. A. Identification of two serine residues involved in agonist activation of the β-adrenergic receptor. J. Biol. Chem. 264, 13572–13578 (1989)
Liapakis, G. et al. The forgotten serine. A critical role for Ser-2035.42 in ligand binding to and activation of the β2-adrenergic receptor. J. Biol. Chem. 275, 37779–37788 (2000)
Warne, T. et al. Structure of a β1-adrenergic G-protein-coupled receptor. Nature 454, 486–491 (2008)
Kobilka, B. K. Amino and carboxyl terminal modifications to facilitate the production and purification of a G protein-coupled receptor. Anal. Biochem. 231, 269–271 (1995)
Wang, Z., Mathias, A., Stavrou, S. & Neville, D. M., Jr A new yeast display vector permitting free scFv amino termini can augment ligand binding affinities. Protein Eng. Des. Sel. 18, 337–343 (2005)
Chao, G. et al. Isolating and engineering human antibodies using yeast surface display. Nature Protocols 1, 755–768 (2006)
Whorton, M. R. et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl Acad. Sci. USA 104, 7682–7687 (2007)
Zou, Y., Weis, W. I. & Kobilka, B. K. N-terminal T4 lysozyme fusion facilitates crystallization of a G protein coupled receptor. PLoS ONE 7, e46039 (2012)
Otwinowski, Z. & Minor, W. in Methods in Enzymology Vol. 276 (ed. Carter, C. W. Jr) 307–326 (Academic, 1997)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Acknowledgements
We thank D. Hilger for critical reading of the manuscript. We acknowledge support from the Stanford Medical Scientist Training Program (A.M. and A.M.R.), the American Heart Association (A.M.), the National Science Foundation (A.C.K.), the Ruth L. Kirschstein National Research Service Award (A.M.R.), National Institutes of Health grants NS02847123 and GM08311806 (B.K.K.), from the Mathers Foundation (B.K.K., W.I.W. and K.C.G.), and from the Howard Hughes Medical Institute (K.C.G.).
Author information
Authors and Affiliations
Contributions
A.M.R. designed and performed yeast display staining and selection experiments, nanobody expression, purification and characterization on yeast and by SPR. A.M. and A.C.K. designed and performed receptor expression, purification, radioligand-binding experiments and crystallography experiments. M.D.E. performed crystallization experiments with adrenaline-bound β2AR under the supervision of A.M. and A.C.K. The manuscript was written by A.M.R., A.M. and A.C.K. W.I.W. supervised structure refinement. K.C.G. and B.K.K. supervised experiments, and B.K.K. supervised manuscript preparation.
Corresponding authors
Ethics declarations
Competing interests
A.M.R., A.M., A.C.K. and B.K.K. have filed a patent application describing methods of generating conformationally selective affinity reagents for transmembrane proteins presented here.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-10, Supplementary Tables 1-3 and an additional reference. (PDF 8205 kb)
Rights and permissions
About this article
Cite this article
Ring, A., Manglik, A., Kruse, A. et al. Adrenaline-activated structure of β2-adrenoceptor stabilized by an engineered nanobody. Nature 502, 575–579 (2013). https://doi.org/10.1038/nature12572
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12572
This article is cited by
-
The chemokine receptor CCR5: multi-faceted hook for HIV-1
Retrovirology (2024)
-
GPR161 structure uncovers the redundant role of sterol-regulated ciliary cAMP signaling in the Hedgehog pathway
Nature Structural & Molecular Biology (2024)
-
Autoregulation of GPCR signalling through the third intracellular loop
Nature (2023)
-
Constrained catecholamines gain β2AR selectivity through allosteric effects on pocket dynamics
Nature Communications (2023)
-
The relaxin receptor RXFP1 signals through a mechanism of autoinhibition
Nature Chemical Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.