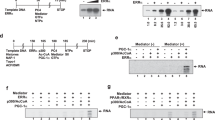
Abstract
The functional importance of gene enhancers in regulated gene expression is well established1,2,3. In addition to widespread transcription of long non-coding RNAs (lncRNAs) in mammalian cells4,5,6, bidirectional ncRNAs are transcribed on enhancers, and are thus referred to as enhancer RNAs (eRNAs)7,8,9. However, it has remained unclear whether these eRNAs are functional or merely a reflection of enhancer activation. Here we report that in human breast cancer cells 17β-oestradiol (E2)-bound oestrogen receptor α (ER-α) causes a global increase in eRNA transcription on enhancers adjacent to E2-upregulated coding genes. These induced eRNAs, as functional transcripts, seem to exert important roles for the observed ligand-dependent induction of target coding genes, increasing the strength of specific enhancer–promoter looping initiated by ER-α binding. Cohesin, present on many ER-α-regulated enhancers even before ligand treatment, apparently contributes to E2-dependent gene activation, at least in part by stabilizing E2/ER-α/eRNA-induced enhancer–promoter looping. Our data indicate that eRNAs are likely to have important functions in many regulated programs of gene transcription.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
04 June 2013
The PDF was corrected to remove two duplicated references from the Methods reference list.
References
Newman, J. J. & Young, R. A. Connecting transcriptional control to chromosome structure and human disease. Cold Spring Harb. Symp. Quant. Biol. 75, 227–235 (2010)
Bulger, M. & Groudine, M. Functional and mechanistic diversity of distal transcription enhancers. Cell 144, 327–339 (2011)
Ong, C. T. & Corces, V. G. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nature Rev. Genet. 12, 283–293 (2011)
Guttman, M. & Rinn, J. L. Modular regulatory principles of large non-coding RNAs. Nature 482, 339–346 (2012)
Wang, K. C. & Chang, H. Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 (2011)
Mercer, T. R., Dinger, M. E. & Mattick, J. S. Long non-coding RNAs: insights into functions. Nature Rev. Genet. 10, 155–159 (2009)
Kim, T.-K. et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187 (2010)
Hah, N. et al. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell 145, 622–634 (2011)
Wang, D. et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474, 390–394 (2011)
Welboren, W. J. et al. ChIP-Seq of ERα and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 28, 1418–1428 (2009)
Carroll, J. S. et al. Genome-wide analysis of estrogen receptor binding sites. Nature Genet. 38, 1289–1297 (2006)
Kwon, Y. S. et al. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor α-binding program on human gene promoters. Proc. Natl Acad. Sci. USA 104, 4852–4857 (2007)
Heintzman, N. D. & Ren, B. Finding distal regulatory elements in the human genome. Curr. Opin. Genet. Dev. 19, 541–549 (2009)
Heintzman, N. D. et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459, 108–112 (2009)
Creyghton, M. P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA 107, 21931–21936 (2010)
Ahlenstiel, C. L. et al. Direct evidence of nuclear Argonaute distribution during transcriptional silencing links the actin cytoskeleton to nuclear RNAi machinery in human cells. Nucleic Acids Res. 40, 1579–1595 (2012)
Mayer, C., Schmitz, K. M., Li, J., Grummt, I. & Santoro, R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell 5, 351–361 (2006)
Wang, K. C. et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124 (2011)
Melo, C. A. et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol. Cell 49, 524–535 (2013)
Lai, F. et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494, 497–501 (2013)
Fullwood, M. J. et al. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462, 58–64 (2009)
Harismendy, O. et al. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signalling response. Nature 470, 264–268 (2011)
Sanyal, A., Lajoie, B. R., Jain, G. & Dekker, J. The long-range interaction landscape of gene promoters. Nature 489, 109–113 (2012)
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009)
Chu, C., Qu, K., Zhong, F. L., Artandi, S. E. & Chang, H. Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 44, 667–678 (2011)
Hadjur, S. et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460, 410–413 (2009)
Kagey, M. H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010)
Schmidt, D. et al. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 20, 578–588 (2010)
Cai, S. & Kohwi-Shigematsu, T. Intranuclear relocalization of matrix binding sites during T cell activation detected by amplified fluorescence in situ hybridization. Methods 19, 394–402 (1999)
Core, L. J., Waterfall, J. J. & Lis, J. T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008)
Abukhdeir, A. M. et al. Physiologic estrogen receptor α signaling in non-tumorigenic human mammary epithelial cells. Breast Cancer Res. Treat. 99, 23–33 (2006)
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010)
Ingolia, N. T. et al. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009)
White, A. K. et al. High-throughput microfluidic single-cell RT-qPCR. Proc. Natl Acad. Sci. USA 108, 13999–14004 (2011)
Zhong, J. F. et al. A microfluidic processor for gene expression profiling of single human embryonic stem cells. Lab Chip 8, 68–74 (2008)
Tsai, M. C. et al. Long non-coding RNA as modular scaffold of histone modification complexes. Science 329, 689–693 (2010)
Rueden, C. T. et al. Visualization approaches for multidimensional biological image data. Biotechniques 43, 31–36 (2007)
Lajoie, B. R. et al. My5C: web tools for chromosome conformation capture studies. Nature Methods 6, 690–691 (2009)
Servant, N. et al. HiTC: exploration of high-throughput ‘C’ experiments. Bioinformatics 28, 2843–2844 (2012)
Stadhouders, R. et al. Dynamic long-range chromatin interactions control Myb proto-oncogene transcription during erythroid development. EMBO J. 31, 986–999 (2011)
Acknowledgements
We thank K. Hutt for help with statistical analyses; M. Ghassemian from the University of California, San Diego (UCSD) Biomolecular/Proteomics Mass Spectrometry Facility for assistance with mass spectrometry; C. Nelson for cell culture assistance; J. Hightower for assistance with figure and manuscript preparation. We thank H. Chang for providing the BoxB, λN–GAL4 constructs. We acknowledge the UCSD Cancer Center Specialized Support Grant P30 CA23100 for confocal microscopy. W.L. and D.N. are supported by Department of Defense (DoD) postdoctoral fellowships, BC110381 and BC103858, respectively. M.G.R. is an investigator with the Howard Hughes Medical Institute. This work was supported by grants DK 039949, DK018477, NS034934, HL065445, CA173903 to C.K.G., and from the DoD.
Author information
Authors and Affiliations
Contributions
M.G.R., W.L., D.N., E.N. and C.K.G. conceived the project. W.L. and D.N. performed most of the experiments reported, with contributions from E.N. and A.Y.C. (FISH). Q.M., B.T. and D.M. performed bioinformatic analyses. Q.M., E.N. and B.T. made equivalent contributions to this study. Additional experiments/methods were contributed by X.S., S.O. and H.-S.K. J.Z. and K.O. assisted in deep-sequencing library preparations and sequencing. W.L., D.N. and M.G.R. wrote the final paper with input from C.K.G.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-9 and Supplementary Tables 1-8. (PDF 3919 kb)
Rights and permissions
About this article
Cite this article
Li, W., Notani, D., Ma, Q. et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520 (2013). https://doi.org/10.1038/nature12210
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12210
This article is cited by
-
Lnc AC016727.1/BACH1/HIF-1 α signal loop promotes the progression of non-small cell lung cancer
Journal of Experimental & Clinical Cancer Research (2023)
-
JUN-induced super-enhancer RNA forms R-loop to promote nasopharyngeal carcinoma metastasis
Cell Death & Disease (2023)
-
The enhancer RNA ADCY10P1 is associated with the progression of ovarian cancer
Journal of Ovarian Research (2022)
-
Enhancer RNA commits osteogenesis via microRNA-3129 expression in human bone marrow-derived mesenchymal stem cells
Inflammation and Regeneration (2022)
-
Discovery of a dual WDR5 and Ikaros PROTAC degrader as an anti-cancer therapeutic
Oncogene (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.