Abstract
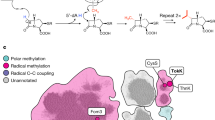
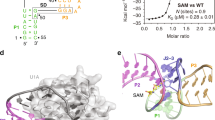
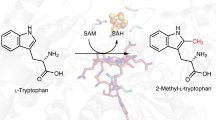
The identification of novel metabolites and the characterization of their biological functions are major challenges in biology. X-ray crystallography can reveal unanticipated ligands that persist through purification and crystallization. These adventitious protein–ligand complexes provide insights into new activities, pathways and regulatory mechanisms. We describe a new metabolite, carboxy-S-adenosyl-l-methionine (Cx-SAM), its biosynthetic pathway and its role in transfer RNA modification. The structure of CmoA, a member of the SAM-dependent methyltransferase superfamily, revealed a ligand consistent with Cx-SAM in the catalytic site. Mechanistic analyses showed an unprecedented role for prephenate as the carboxyl donor and the involvement of a unique ylide intermediate as the carboxyl acceptor in the CmoA-mediated conversion of SAM to Cx-SAM. A second member of the SAM-dependent methyltransferase superfamily, CmoB, recognizes Cx-SAM and acts as a carboxymethyltransferase to convert 5-hydroxyuridine into 5-oxyacetyl uridine at the wobble position of multiple tRNAs in Gram-negative bacteria1, resulting in expanded codon-recognition properties2,3. CmoA and CmoB represent the first documented synthase and transferase for Cx-SAM. These findings reveal new functional diversity in the SAM-dependent methyltransferase superfamily and expand the metabolic and biological contributions of SAM-based biochemistry. These discoveries highlight the value of structural genomics approaches in identifying ligands within the context of their physiologically relevant macromolecular binding partners, and in revealing their functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Czerwoniec, A. et al. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 37, D118–D121 (2009)
Nasvall, S. J., Chen, P. & Bjork, G. R. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA 10, 1662–1673 (2004)
Näsvall, S. J., Chen, P. & Bjork, G. R. The wobble hypothesis revisited: uridine-5-oxyacetic acid is critical for reading of G-ending codons. RNA 13, 2151–2164 (2007)
Weixlbaumer, A. et al. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nature Struct. Mol. Biol. 14, 498–502 (2007)
Björk, G. R. A novel link between the biosynthesis of aromatic amino acids and transfer RNA modification in Escherichia coli. J. Mol. Biol. 140, 391–410 (1980)
Hagervall, T. G., Jonsson, Y. H., Edmonds, C. G., McCloskey, J. A. & Bjork, G. R. Chorismic acid, a key metabolite in modification of tRNA. J. Bacteriol. 172, 252–259 (1990)
Lim, K. et al. Crystal structure of YecO from Haemophilus influenzae (HI0319) reveals a methyltransferase fold and a bound S-adenosylhomocysteine. Proteins 45, 397–407 (2001)
Van Vleet, J., Kleeb, A., Kast, P., Hilvert, D. & Cleland, W. W. 13C isotope effect on the reaction catalyzed by prephenate dehydratase. Biochim. Biophys. Acta 1804, 752–754 (2010)
Horowitz, S., Yesselman, J. D., Al-Hashimi, H. M. & Trievel, R. C. Direct evidence for methyl group coordination by carbon-oxygen hydrogen bonds in the lysine methyltransferase SET7/9. J. Biol. Chem. 286, 18658–18663 (2011)
Crosby, J. & Stirling, C. J. M. Elimination and addition reactions. Part XIX. Elimination of phenoxide from β-substituted ethyl phenyl ethers: the nature of activation in 1,2-elimination. J. Chemic. Soc. B 671–679 (1970)
Bordwell, F. G. Equilibrium acidities in dimethyl sulfoxide solution. Acc. Chem. Res. 21, 456–463 (1988)
Arrowsmith, C. H., Bountra, C., Fish, P. V., Lee, K. & Schapira, M. Epigenetic protein families: a new frontier for drug discovery. Nature Rev. Drug Discov. 11, 384–400 (2012)
Cedar, H. & Bergman, Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature Rev. Genet. 10, 295–304 (2009)
Luka, Z., Mudd, S. H. & Wagner, C. Glycine N-methyltransferase and regulation of S-adenosylmethionine levels. J. Biol. Chem. 284, 22507–22511 (2009)
Vévodová, J. et al. Structure/function studies on a S-adenosyl-l-methionine-dependent uroporphyrinogen III C methyltransferase (SUMT), a key regulatory enzyme of tetrapyrrole biosynthesis. J. Mol. Biol. 344, 419–433 (2004)
Kowtoniuk, W. E., Shen, Y., Heemstra, J. M., Agarwal, I. & Liu, D. R. A chemical screen for biological small molecule-RNA conjugates reveals CoA-linked RNA. Proc. Natl Acad. Sci. USA 106, 7768–7773 (2009)
Dalhoff, C., Lukinavicius, G., Klimasauskas, S. & Weinhold, E. Direct transfer of extended groups from synthetic cofactors by DNA methyltransferases. Nature Chem. Biol. 2, 31–32 (2006)
Dalhoff, C., Lukinavicius, G., Klimasauskas, S. & Weinhold, E. Synthesis of S-adenosyl-l-methionine analogs and their use for sequence-specific transalkylation of DNA by methyltransferases. Nature Protocols 1, 1879–1886 (2006)
Binda, O. et al. A chemical method for labeling lysine methyltransferase substrates. ChemBioChem 12, 330–334 (2011)
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D 62, 859–866 (2006)
Lebedev, A. A., Vagin, A. A. & Murshudov, G. N. Model preparation in MOLREP and examples of model improvement using X-ray data. Acta Crystallogr. D 64, 33–39 (2008)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Dopheide, T. A., Crewther, P. & Davidson, B. E. Chorismate mutase-prephenate dehydratase from Escherichia coli K-12. II. Kinetic properties. J. Biol. Chem. 247, 4447–4452 (1972)
Lorenz, M. A., Burant, C. F. & Kennedy, R. T. Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Anal. Chem. 83, 3406–3414 (2011)
Kalyanaraman, C., Bernacki, K. & Jacobson, M. P. Virtual screening against highly charged active sites: identifying substrates of alpha-beta barrel enzymes. Biochemistry 44, 2059–2071 (2005)
Gibson, F. Chorismic acid: purification and some chemical and physical studies. Biochem. J. 90, 256–261 (1964)
Parker, J. B. & Walsh, C. T. Olefin isomerization regiochemistries during tandem action of BacA and BacB on prephenate in bacilysin biosynthesis. Biochemistry 51, 3241–3251 (2012)
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402 (1997)
Punta, M. et al. The Pfam protein families database. Nucleic Acids Res 40, D290–D301 (2008)
Pegg, S. C. et al. Leveraging enzyme structure-function relationships for functional inference and experimental design: the structure-function linkage database. Biochemistry 45, 2545–2555 (2006)
Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P. L. & Ideker, T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 (2012)
Uniprot Consortium Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 40, D71–D75 (2012)
Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J. & Sayers, E. W. GenBank. Nucleic Acids Res. 37, D26–D31 (2009)
Sayers, E. W. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 37, D5–D15 (2009)
Edgar, R. C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (2004)
Acknowledgements
We thank J. Parker and C. T. Walsh for providing the Aerobacter aerogenes 62-1 strain. We are indebted to V. Schramm and J. Gerlt for critical discussions and reading of the manuscript. This work was supported by US National Institutes of Health grants GM094662 (to S.C.A.), GM093342 (to S.C.A., M.P.J. and P.C.B.) and the Albert Einstein Cancer Center. This publication was made possible by the Center for Synchrotron Biosciences grant P30-EB-009998 from the National Institute of Biomedical Imaging and Bioengineering (NIBIB). Use of the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract no. DE-AC02-98CH10886.
Author information
Authors and Affiliations
Contributions
J.K. carried out cloning, protein purification, crystallography, and functional assays. H.X. performed mass-spectrometry analysis of the in vitro assay. Y.-S.L. carried out LC–MS analysis of the CmoA-bound ligand and chemical synthesis of Cx-SAM. X.T. performed the NMR experiments. N.F.A.-O. carried out thermal denaturation studies. C.K. and M.P.J. performed computational modelling. S.B. and P.C.B. carried out the bioinformatics analysis. J.B.B. and Y.P. assisted in crystallographic validation and analysed crystallographic ligand-binding results. J.K. and S.C.A. designed the study, analysed the data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Table 1, Supplementary Figures 1-14 and Supplementary References. (PDF 5390 kb)
Rights and permissions
About this article
Cite this article
Kim, J., Xiao, H., Bonanno, J. et al. Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function. Nature 498, 123–126 (2013). https://doi.org/10.1038/nature12180
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12180
This article is cited by
-
Biotechnological applications of S-adenosyl-methionine-dependent methyltransferases for natural products biosynthesis and diversification
Bioresources and Bioprocessing (2021)
-
Challenging nature’s preference for methylation
Nature Chemistry (2020)
-
Structural and biochemical characterization of Rv0187, an O-methyltransferase from Mycobacterium tuberculosis
Scientific Reports (2019)
-
Dual pathways of tRNA hydroxylation ensure efficient translation by expanding decoding capability
Nature Communications (2019)
-
Maintenance of protein synthesis reading frame by EF-P and m1G37-tRNA
Nature Communications (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.