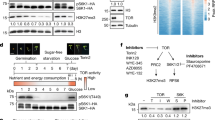
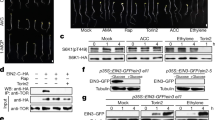
Abstract
Meristems encompass stem/progenitor cells that sustain postembryonic growth of all plant organs. How meristems are activated and sustained by nutrient signalling remains enigmatic in photosynthetic plants. Combining chemical manipulations and chemical genetics at the photoautotrophic transition checkpoint, we reveal that shoot photosynthesis-derived glucose drives target-of-rapamycin (TOR) signalling relays through glycolysis and mitochondrial bioenergetics to control root meristem activation, which is decoupled from direct glucose sensing, growth-hormone signalling and stem-cell maintenance. Surprisingly, glucose–TOR signalling dictates transcriptional reprogramming of remarkable gene sets involved in central and secondary metabolism, cell cycle, transcription, signalling, transport and protein folding. Systems, cellular and genetic analyses uncover TOR phosphorylation of E2Fa transcription factor for an unconventional activation of S-phase genes, and glucose-signalling defects in e2fa root meristems. Our findings establish pivotal roles of glucose–TOR signalling in unprecedented transcriptional networks wiring central metabolism and biosynthesis for energy and biomass production, and integrating localized stem/progenitor-cell proliferation through inter-organ nutrient coordination to control developmental transition and growth.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Chen, L. Q. et al. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335, 207–211 (2012)
Aichinger, E., Kornet, N., Friedrich, T. & Laux, T. Plant stem cell niches. Annu. Rev. Plant Biol. 63, 615–636 (2012)
Baena-González, E. & Sheen, J. Convergent energy and stress signaling. Trends Plant Sci. 13, 474–482 (2008)
Robaglia, C., Thomas, M. & Meyer, C. Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr. Opin. Plant Biol. 15, 301–307 (2012)
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012)
Dowling, R. J. et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328, 1172–1176 (2010)
Hsu, P. P. et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 (2011)
Yu, Y. et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332, 1322–1326 (2011)
Hsieh, A. C. et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 (2012)
Moreau, M. et al. Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 24, 463–481 (2012)
Moore, B. et al. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300, 332–336 (2003)
Graham, I. A. Seed storage oil mobilization. Annu. Rev. Plant Biol. 59, 115–142 (2008)
Xiong, Y. & Sheen, J. Rapamycin and glucose-target of rapamycin (TOR) protein signaling in plants. J. Biol. Chem. 287, 2836–2842 (2012)
Sanz, L. et al. The Arabidopsis D-type cyclin CYCD2;1 and the inhibitor ICK2/KRP2 modulate auxin-induced lateral root formation. Plant Cell 23, 641–660 (2011)
Müller, B. & Sheen, J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453, 1094–1097 (2008)
Zhang, Z. W. et al. The plastid hexokinase pHXK: a node of convergence for sugar and plastid signals in Arabidopsis. FEBS Lett. 584, 3573–3579 (2010)
Kotogány, E., Dudits, D., Horvath, G. V. & Ayaydin, F. A rapid and robust assay for detection of S-phase cell cycle progression in plant cells and tissues by using ethynyl deoxyuridine. Plant Methods 6, 5 (2010)
Chaudhuri, B. et al. Protonophore- and pH-insensitive glucose and sucrose accumulation detected by FRET nanosensors in Arabidopsis root tips. Plant J. 56, 948–962 (2008)
Hirose, N. et al. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 59, 75–83 (2008)
Tsukagoshi, H., Busch, W. & Benfey, P. N. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143, 606–616 (2010)
Moubayidin, L. et al. The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr. Biol. 20, 1138–1143 (2010)
Boudsocq, M. et al. Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464, 418–422 (2010)
Gonzali, S. et al. Identification of sugar-modulated genes and evidence for in vivo sugar sensing in Arabidopsis. J. Plant Res. 119, 115–123 (2006)
Li, Y. et al. Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Res. 16, 414–427 (2006)
Bläsing, O. E. et al. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17, 3257–3281 (2005)
Matsuzaki, Y., Ogawa-Ohnishi, M., Mori, A. & Matsubayashi, Y. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329, 1065–1067 (2010)
Vernoux, T. et al. The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 gene defines a glutathione-dependent pathway involved in initiation and maintenance of cell division during postembryonic root development. Plant Cell 12, 97–110 (2000)
Thimm, O. et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939 (2004)
Urban, J. et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26, 663–674 (2007)
Hardwick, J. S., Kuruvilla, F. G., Tong, J. K., Shamji, A. F. & Schreiber, S. L. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl Acad. Sci. USA 96, 14866–14870 (1999)
Düvel, K. et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 39, 171–183 (2010)
Baena-González, E., Rolland, F., Thevelein, J. M. & Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942 (2007)
Menges, M., Hennig, L., Gruissem, W. & Murray, J. A. Genome-wide gene expression in an Arabidopsis cell suspension. Plant Mol. Biol. 53, 423–442 (2003)
de Jager, S. M. et al. Dissecting regulatory pathways of G1/S control in Arabidopsis: common and distinct targets of CYCD3;1, E2Fa and E2Fc. Plant Mol. Biol. 71, 345–365 (2009)
Naouar, N. et al. Quantitative RNA expression analysis with Affymetrix Tiling 1.0R arrays identifies new E2F target genes. Plant J. 57, 184–194 (2009)
Vandepoele, K. et al. Genome-wide identification of potential plant E2F target genes. Plant Physiol. 139, 316–328 (2005)
Liu, Q. et al. Kinome-wide selectivity profiling of ATP-competitive mammalian target of rapamycin (mTOR) inhibitors and characterization of their binding kinetics. J. Biol. Chem. 287, 9742–9752 (2012)
Magyar, Z. et al. Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J. 31, 1480–1493 (2012)
Cunningham, J. T. et al. mTOR controls mitochondrial oxidative function through a YY1-PGC-1α transcriptional complex. Nature 450, 736–740 (2007)
Dello Ioio, R. et al. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17, 678–682 (2007)
Fu, X. & Harberd, N. P. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421, 740–743 (2003)
González-Garcia, M. P. et al. Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138, 849–859 (2011)
Acknowledgements
We thank N. Dai and J. Avruch for S6K antibodies and advice, J. L. Celenza for stimulating discussion, M. D. Curtis and Y. J. Niu for the oestradiol-inducible binary vector, L. Li and J. Bush for seeds and plants, B. Müller for TCS::GFP, J. Friml for DR5::GFP, N. S. Gray and D. M. Sabatini for torin1, and J. F. Li, H. Lee and M. Ramon for critical reading of the manuscript. Y.X. is supported by the MGH Tosteson Postdoctoral Fellowship. C.X. is supported by Chinese Academy of Sciences (KSCX3-YW-N-007). The Research is supported by the NSF, NIH and WJC Special Project (PJ009106) RDA-Korea to J.S.
Author information
Authors and Affiliations
Contributions
Y.X. and J.S. initiated the project and designed the experiments; Y.X. carried out most of the experiments; L.L. and Y.X. conducted quantitative ChIP-PCR analyses; Y.X., M.M. and J.S. analysed the microarray data. C.X. isolated the e2fa mutant. Q.H. generated PLT::GFP and WOX5::GFP transgenic lines. Y.X., M.M. and J.S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contain Supplementary Figures 1-22, legends for Supplementary Tables 1-2, 5-7 (see separate files for tables), Supplementary Tables 3-4, 8-11, Supplementary Methods and Supplementary References. (PDF 2819 kb)
Supplementary Table 1
This file contains the Glucose-TOR target genes – see Supplementary Information file for full legend. (XLSX 259 kb)
Supplementary Table 2
This file contains the Novel glucose regulated genes – see Supplementary Information file for full legend. (XLSX 47 kb)
Supplementary Table 5
This file contains the Glucose-TOR target gene list (P value <0.01) – see Supplementary Information file for full legend. (XLSX 426 kb)
Supplementary Table 6
This file contains the Glucose-TOR target genes involved in cell cycle – see Supplementary Information file for full legend. (XLSX 76 kb)
Supplementary Table 7
This file contains the Putative E2Fa target genes – see Supplementary Information file for full legend. (XLSX 367 kb)
Rights and permissions
About this article
Cite this article
Xiong, Y., McCormack, M., Li, L. et al. Glucose–TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186 (2013). https://doi.org/10.1038/nature12030
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12030
This article is cited by
-
The proteomics and metabolomics studies of GZU001 on promoting the Merisis of maize (Zea mays L.) roots
BMC Plant Biology (2023)
-
Unlocking the potentials of cyanobacterial photosynthesis for directly converting carbon dioxide into glucose
Nature Communications (2023)
-
High Sugar Concentration Inhibits TOR Signaling Pathway in Arabidopsis thaliana
Journal of Plant Growth Regulation (2023)
-
Regulation of early seedling establishment and root development in Arabidopsis thaliana by light and carbohydrates
Planta (2023)
-
Mining genes regulating root system architecture in maize based on data integration analysis
Theoretical and Applied Genetics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.