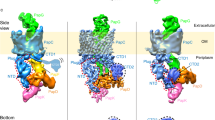
Abstract
Type 1 pili, produced by uropathogenic Escherichia coli, are multisubunit fibres crucial in recognition of and adhesion to host tissues1. During pilus biogenesis, subunits are recruited to an outer membrane assembly platform, the FimD usher, which catalyses their polymerization and mediates pilus secretion2. The recent determination of the crystal structure of an initiation complex provided insight into the initiation step of pilus biogenesis resulting in pore activation, but very little is known about the elongation steps that follow3. Here, to address this question, we determine the structure of an elongation complex in which the tip complex assembly composed of FimC, FimF, FimG and FimH passes through FimD. This structure demonstrates the conformational changes required to prevent backsliding of the nascent pilus through the FimD pore and also reveals unexpected properties of the usher pore. We show that the circular binding interface between the pore lumen and the folded substrate participates in transport by defining a low-energy pathway along which the nascent pilus polymer is guided during secretion.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Waksman, G. & Hultgren, S. J. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nature Rev. Microbiol. 7, 765–774 (2009)
Nishiyama, M., Ishikawa, T., Rechsteiner, H. & Glockshuber, R. Reconstitution of pilus assembly reveals a bacterial outer membrane catalyst. Science 320, 376–379 (2008)
Phan, G. et al. Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate. Nature 474, 49–53 (2011)
Sauer, F. G. et al. Structural basis of chaperone function and pilus biogenesis. Science 285, 1058–1061 (1999)
Sauer, F. G., Pinkner, J. S., Waksman, G. & Hultgren, S. J. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell 111, 543–551 (2002)
Choudhury, D. et al. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285, 1061–1066 (1999)
Zavialov, A. V. et al. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell 113, 587–596 (2003)
Vetsch, M. et al. Pilus chaperones represent a new type of protein-folding catalyst. Nature 431, 329–333 (2004)
Le Trong, I. et al. Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like β-sheet twisting. Cell 141, 645–655 (2010)
Das, R. & Baker, D. Macromolecular modeling with Rosetta. Annu. Rev. Biochem. 77, 363–382 (2008)
Leaver-Fay, A. et al. Rosetta3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487, 545–574 (2011)
Huang, Y., Smith, B. S., Chen, L. X., Baxter, R. H. & Deisenhofer, J. Insights into pilus assembly and secretion from the structure and functional characterization of usher PapC. Proc. Natl Acad. Sci. USA 106, 7403–7407 (2009)
Remaut, H. et al. Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Cell 133, 640–652 (2008)
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010)
Karplus, P. A. & Diederichs, K. Linking crystallographic model and data quality. Science 336, 1030–1033 (2012)
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006)
McCoy, A. J. et al. PHASER crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of COOT. Acta Crystallogr. D 66, 486–501 (2010)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011)
Schröder, G. F., Levitt, M. & Brunger, A. T. Super-resolution biomolecular crystallography with low-resolution data. Nature 464, 1218–1222 (2010)
Brunger, A. T. Version 1.2 of the Crystallography and NMR system. Nature Protocols 2, 2728–2733 (2007)
O’Donovan, D. J. et al. A grid-enabled web service for low-resolution crystal structure refinement. Acta Crystallogr. D 68, 261–267 (2012)
DiMaio, F., Tyka, M. D., Baker, M. L., Chiu, W. & Baker, D. Refinement of protein structures into low-resolution density maps using Rosetta. J. Mol. Biol. 392, 181–190 (2009)
Fleishman, S. J. et al. RosettaScripts: a scripting language interface to the Rosetta macromolecular modeling suite. PLoS ONE 6, e20161 (2011)
Feig, M., Karanicolas, J. & Brooks, C. L., III MMTSB tool set: enhanced sampling and multiscale modeling methods for applications in structural biology. J. Mol. Graph. Model. 22, 377–395 (2004)
Acknowledgements
This work was funded by Medical Research Council grant 85602 to G.W. D.B. and E.P. are supported by grant P41 GM103533 from the National Institute of General Medical Studies at the US National Institutes of Health (NIH). S.J.H. was supported by grant AI029549 from the National Institute of Allergy and Infectious Disease at the NIH. We thank the staff of beamline ID23-1 at the European Synchrotron Radiation Facility, the staff of beamline IO2 at the Diamond Light source and A. Cole for technical assistance during data collection.
Author information
Authors and Affiliations
Contributions
S.G. and E.P. carried out the crystallographic and computational work, respectively. D.B. and G.W. supervised the work. S.G., E.P., D.B. and G.W. analysed the data. S.G., E.P., S.J.H., D.B. and G.W. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Tables 1-2, Supplementary Figures 1-16 and Supplementary References. (PDF 23699 kb)
Rights and permissions
About this article
Cite this article
Geibel, S., Procko, E., Hultgren, S. et al. Structural and energetic basis of folded-protein transport by the FimD usher. Nature 496, 243–246 (2013). https://doi.org/10.1038/nature12007
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12007
This article is cited by
-
The assembly platform FimD is required to obtain the most stable quaternary structure of type 1 pili
Nature Communications (2024)
-
Stochastic chain termination in bacterial pilus assembly
Nature Communications (2023)
-
Classical chaperone-usher (CU) adhesive fimbriome: uropathogenic Escherichia coli (UPEC) and urinary tract infections (UTIs)
Folia Microbiologica (2020)
-
Characterization of H/D exchange in type 1 pili by proton-detected solid-state NMR and molecular dynamics simulations
Journal of Biomolecular NMR (2019)
-
Structural basis for usher activation and intramolecular subunit transfer in P pilus biogenesis in Escherichia coli
Nature Microbiology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.