Abstract
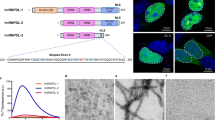
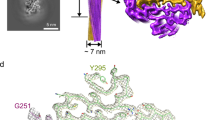
Algorithms designed to identify canonical yeast prions predict that around 250 human proteins, including several RNA-binding proteins associated with neurodegenerative disease, harbour a distinctive prion-like domain (PrLD) enriched in uncharged polar amino acids and glycine. PrLDs in RNA-binding proteins are essential for the assembly of ribonucleoprotein granules. However, the interplay between human PrLD function and disease is not understood. Here we define pathogenic mutations in PrLDs of heterogeneous nuclear ribonucleoproteins (hnRNPs) A2B1 and A1 in families with inherited degeneration affecting muscle, brain, motor neuron and bone, and in one case of familial amyotrophic lateral sclerosis. Wild-type hnRNPA2 (the most abundant isoform of hnRNPA2B1) and hnRNPA1 show an intrinsic tendency to assemble into self-seeding fibrils, which is exacerbated by the disease mutations. Indeed, the pathogenic mutations strengthen a ‘steric zipper’ motif in the PrLD, which accelerates the formation of self-seeding fibrils that cross-seed polymerization of wild-type hnRNP. Notably, the disease mutations promote excess incorporation of hnRNPA2 and hnRNPA1 into stress granules and drive the formation of cytoplasmic inclusions in animal models that recapitulate the human pathology. Thus, dysregulated polymerization caused by a potent mutant steric zipper motif in a PrLD can initiate degenerative disease. Related proteins with PrLDs should therefore be considered candidates for initiating and perhaps propagating proteinopathies of muscle, brain, motor neuron and bone.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Nalbandian, A. et al. The multiple faces of valosin-containing protein-associated diseases: inclusion body myopathy with Paget’s disease of bone, frontotemporal dementia, and amyotrophic lateral sclerosis. J. Mol. Neurosci. 45, 522–531 (2011)
Johnson, J. O. et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 68, 857–864 (2010)
Watts, G. D. et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nature Genet. 36, 377–381 (2004)
Neumann, M., Tolnay, M. & Mackenzie, I. R. The molecular basis of frontotemporal dementia. Expert Rev. Mol. Med. 11, e23 (2009)
Shi, Z. et al. Characterization of the Asian myopathy patients with VCP mutations. Eur. J. Neurol. 19, 501–509 (2012)
Chung, P. Y. et al. Indications for a genetic association of a VCP polymorphism with the pathogenesis of sporadic Paget’s disease of bone, but not for TNFSF11 (RANKL) and IL-6 polymorphisms. Mol. Genet. Metab. 103, 287–292 (2011)
Kottlors, M. et al. Late-onset autosomal dominant limb girdle muscular dystrophy and Paget’s disease of bone unlinked to the VCP gene locus. J. Neurol. Sci. 291, 79–85 (2010)
Buratti, E. et al. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 280, 37572–37584 (2005)
Ritson, G. P. et al. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J. Neurosci. 30, 7729–7739 (2010)
Iwahashi, C. K. et al. Protein composition of the intranuclear inclusions of FXTAS. Brain 129, 256–271 (2006)
Sofola, O. A. et al. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron 55, 565–571 (2007)
Jin, P. et al. Pur α binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron 55, 556–564 (2007)
Salajegheh, M. et al. Sarcoplasmic redistribution of nuclear TDP-43 in inclusion body myositis. Muscle Nerve 40, 19–31 (2009)
Alberti, S., Halfmann, R., King, O., Kapila, A. & Lindquist, S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137, 146–158 (2009)
Toombs, J. A., McCarty, B. R. & Ross, E. D. Compositional determinants of prion formation in yeast. Mol. Cell. Biol. 30, 319–332 (2010)
Goldschmidt, L., Teng, P. K., Riek, R. & Eisenberg, D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl Acad. Sci. USA 107, 3487–3492 (2010)
Teng, P. K. & Eisenberg, D. Short protein segments can drive a non-fibrillizing protein into the amyloid state. Protein Eng. Des. Sel. 22, 531–536 (2009)
Li, L. & Lindquist, S. Creating a protein-based element of inheritance. Science 287, 661–664 (2000)
Kato, M. et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 (2012)
Wolozin, B. Regulated protein aggregation: stress granules and neurodegeneration. Mol. Neurodegener. 7, 56 (2012)
Buchan, J. R. & Parker, R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941 (2009)
Weber, S. C. & Brangwynne, C. P. Getting RNA and protein in phase. Cell 149, 1188–1191 (2012)
Neumann, M. et al. FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain 134, 2595–2609 (2011)
King, O. D., Gitler, A. D. & Shorter, J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 1462, 61–80 (2012)
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010)
Katoh, K., Asimenos, G. & Toh, H. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 537, 39–64 (2009)
Goode, M. G. & Rodrigo, A. G. SQUINT: a multiple alignment program and editor. Bioinformatics 23, 1553–1555 (2007)
Zwickl, D. J. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets Under the Maximum Likelihood Criterion. PhD thesis, Univ. Texas at Austin. (2006)
Johnson, B. S., McCaffery, J. M., Lindquist, S. & Gitler, A. D. A yeast TDP-43 proteinopathy model: exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc. Natl Acad. Sci. USA 105, 6439–6444 (2008)
Johnson, B. S. et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 284, 20329–20339 (2009)
Sun, Z. et al. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 9, e1000614 (2011)
Alberti, S., Gitler, A. D. & Lindquist, S. A suite of Gateway® cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae . Yeast 24, 913–919 (2007)
Ross, E. D., Edskes, H. K., Terry, M. J. & Wickner, R. B. Primary sequence independence for prion formation. Proc. Natl Acad. Sci. USA 102, 12825–12830 (2005)
Song, Y. et al. Role for Hsp70 chaperone in Saccharomyces cerevisiae prion seed replication. Eukaryot. Cell 4, 289–297 (2005)
Ross, C. D., McCarty, B. M., Hamilton, M., Ben-Hur, A. & Ross, E. D. A promiscuous prion: efficient induction of [URE3] prion formation by heterologous prion domains. Genetics 183, 929–940 (2009)
Guthrie, C. & Fink, G. R. Methods in Ezymology: Guide to Yeast Genetics and Molecular and Cell Biology 169 (Academic, 2002)
Ito, H., Fukuda, Y., Murata, K. & Kimura, A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168 (1983)
Ross, E. D., Edskes, H. K., Terry, M. J. & Wickner, R. B. Primary sequence independence for prion formation. Proc. Natl Acad. Sci. USA 102, 12825–12830 (2005)
Acknowledgements
We thank the patients whose participation made this work possible. We thank the St Jude Pediatric Cancer Genome Project and J. Zhang in particular for providing access to control sequencing data. We thank C. Gellera, B. Baloh, M. Harms, S. Krause, G. Dreyfuss and T. Cundy for sharing reagents. We thank S. Donkervoort and S. Mumm for coordinating samples, and A. Taylor for editorial assistance. J.P.T. was supported by ALSAC, the Packard Foundation and the National Institutes of Health (NIH) (NS053825); J.P.T. and M.B. were supported by the ALS Association; J.Q.T. was supported by the NIH (AG032953); J.S. was supported by the NIH (DP2OD002177 and NS067354) and the Ellison Medical Foundation; E.D.R. was supported by the National Science Foundation (MCB-1023771). C.C.W. was supported by the NIH (AG031867).
Author information
Authors and Affiliations
Contributions
H.J.K., N.C.K., E.D.R., C.C.W., J.S. and J.P.T. designed experiments. H.J.K., N.C.K., E.A.S., J.Moore, Z.D., K.S.M., B.F., S.L., A.M., A.P.K., Y.R.L. and A.F.F. performed the experiments. K.B.B., A.M.W., R.R., J.L.P., S.A.G., J.Q.T., B.N.S., S.T., A.-S.G., J.Miller, C.E.S., M.K., J.K., A.P., M.B. and V.E.K. provided patient clinical material, clinical evaluation, or evaluation of patient clinical material. H.J.K., N.C.K., Y.-D.W., R.C., B.J.T., A.D.G., O.D.K., E.D.R., J.S. and J.P.T. contributed to data analysis. E.D.R., O.D.K. and C.C.W. contributed to manuscript preparation. H.J.K., J.S. and J.P.T. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-18, Supplementary Tables 1-3 and additional references. (PDF 14474 kb)
Rights and permissions
About this article
Cite this article
Kim, H., Kim, N., Wang, YD. et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473 (2013). https://doi.org/10.1038/nature11922
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11922
This article is cited by
-
Nuclear-import receptors as gatekeepers of pathological phase transitions in ALS/FTD
Molecular Neurodegeneration (2024)
-
Physiology and pharmacological targeting of phase separation
Journal of Biomedical Science (2024)
-
A distant global control region is essential for normal expression of anterior HOXA genes during mouse and human craniofacial development
Nature Communications (2024)
-
Physiological and pathological effects of phase separation in the central nervous system
Journal of Molecular Medicine (2024)
-
The Roles of hnRNP Family in the Brain and Brain-Related Disorders
Molecular Neurobiology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.