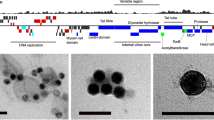
Abstract
Several reports proposed that the extraordinary dominance of the SAR11 bacterial clade in ocean ecosystems could be a consequence of unusual mechanisms of resistance to bacteriophage infection, including ‘cryptic escape’ through reduced cell size1 and/or K-strategist defence specialism2. Alternatively, the evolution of high surface-to-volume ratios coupled with minimal genomes containing high-affinity transporters enables unusually efficient metabolism for oxidizing dissolved organic matter in the world’s oceans that could support vast population sizes despite phage susceptibility. These ideas are important for understanding plankton ecology because they emphasize the potentially important role of top-down mechanisms in predation, thus determining the size of SAR11 populations and their concomitant role in biogeochemical cycling. Here we report the isolation of diverse SAR11 viruses belonging to two virus families in culture, for which we propose the name ‘pelagiphage’, after their host. Notably, the pelagiphage genomes were highly represented in marine viral metagenomes, demonstrating their importance in nature. One of the new phages, HTVC010P, represents a new podovirus subfamily more abundant than any seen previously, in all data sets tested, and may represent one of the most abundant virus subfamilies in the biosphere. This discovery disproves the theory that SAR11 cells are immune to viral predation and is consistent with the interpretation that the success of this highly abundant microbial clade is the result of successfully evolved adaptation to resource competition.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
Primary accessions
GenBank/EMBL/DDBJ
Data deposits
Viral genome sequences and annotations have been deposited at GenBank/EMBL/DDBJ under accession codes: KC465898 (HTVC10P), KC465899 (HTVC008M), KC465900 (HTVC011P) and KC465901 (HTVC019P). Scripts and data used to generate the figures in this manuscript are available at http://giovannonilab.science.oregonstate.edu/publications.
References
Yooseph, S. et al. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature 468, 60–66 (2010)
Suttle, C. A. Marine viruses — major players in the global ecosystem. Nature Rev. Microbiol. 5, 801–812 (2007)
Nagasaki, K. & Bratbak, G. In Manual of Aquatic Viral Ecology Limnol . Oceanogr. 92–101 (2010)
Lavigne, R., Seto, D., Mahadevan, P., Ackermann, H.-W. & Kropinski, A. M. Unifying classical and molecular taxonomic classification: analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol. 159, 406–414 (2008)
Ignacio-Espinoza, J. C. & Sullivan, M. B. Phylogenomics of T4 cyanophages: lateral gene transfer in the ‘core’ and origins of host genes. Environ. Microbiol. 14, 2113–2126 (2012)
Hurwitz, B. L. & Sullivan, M. B. The Pacific Ocean virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS ONE (in the press). (2013)
Duhaime, M. B., Deng, L., Poulos, B. T. & Sullivan, M. B. Towards quantitative metagenomics of wild viruses and other ultra-low concentration DNA samples: a rigorous assessment and optimization of the linker amplification method. Environ. Microbiol. 14, 2526–2537 (2012)
Angly, F. E. et al. The marine viromes of four oceanic regions. PLoS Biol. 4, e368 (2006)
López-Bueno, A. et al. High diversity of the viral community from an Antarctic lake. Science 326, 858–861 (2009)
Ghai, R. et al. Metagenome of the Mediterranean deep chlorophyll maximum studied by direct and fosmid library 454 pyrosequencing. ISME J. 4, 1154–1166 (2010)
Wilhelm, L. J., Tripp, H. J., Givan, S. A., Smith, D. P. & Giovannoni, S. J. Natural variation in SAR11 marine bacterioplankton genomes inferred from metagenomic data. Biol. Direct 2, 27 (2007)
Grote, J. et al. Streamlining and core genome conservation among highly divergent members of the SAR11 clade. mBio 3, http://dx.doi.org/10.1128/mBio.00252-12 (2012)
Rodriguez-Valera, F. et al. Explaining microbial population genomics through phage predation. Nature Rev. Microbiol. 7, 828–836 (2009)
Avrani, S., Wurtzel, O., Sharon, I., Sorek, R. & Lindell, D. Genomic island variability facilitates Prochlorococcus–virus coexistence. Nature 474, 604–608 (2011)
Wang, L. et al. DNA phosphorothioation is widespread and quantized in bacterial genomes. Proc. Natl Acad. Sci. USA 108, 2963–2968 (2011)
Thingstad, T. F. & Lignell, R. Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat. Microb. Ecol. 13, 19–27 (1997)
Chow, C.-E. T. & Fuhrman, J. A. Seasonality and monthly dynamics of marine myovirus communities. Environ. Microbiol. 14, 2171–2183 (2012)
Marston, M. F. et al. Rapid diversification of coevolving marine Synechococcus and a virus. Proc. Natl Acad. Sci. USA 109, 4544–4549 (2012)
Fuhrman, J. A. & Schwalbach, M. Viral influence on aquatic bacterial communities. Biol. Bull. 204, 192–195 (2003)
Parsons, R. J., Breitbart, M., Lomas, M. W. & Carlson, C. A. Ocean time-series reveals recurring seasonal patterns of virioplankton dynamics in the northwestern Sargasso Sea. ISME J. 6, 273–284 (2012)
Rappé, M. S., Connon, S. A., Vergin, K. L. & Giovannoni, S. J. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418, 630–633 (2002)
Sullivan, M. B., Waterbury, J. B. & Chisholm, S. W. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus . Nature 424, 1047–1051 (2003)
Sullivan, M. B., Coleman, M., Weigele, P., Rohwer, F. & Chisholm, S. W. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3, e144 (2005)
Sullivan, M. B. et al. The genome and structural proteome of an ocean siphovirus: a new window into the cyanobacterial ‘mobilome’. Environ. Microbiol. 11, 2935–2951 (2009)
Sullivan, M. B. et al. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ. Microbiol. 12, 3035–3056 (2010)
Malmstrom, R. R., Cottrell, M. T., Elifantz, H. & Kirchman, D. L. Biomass production and assimilation of dissolved organic matter by SAR11 bacteria in the Northwest Atlantic Ocean. Appl. Environ. Microbiol. 71, 2979–2986 (2005)
Kunin, V. et al. A bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res. 18, 293–297 (2008)
Vergin, K. L. et al. High intraspecific recombination rate in a native population of Candidatus Pelagibacter ubique (SAR11). Environ. Microbiol. 9, 2430–2440 (2007)
Vos, M. & Didelot, X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 3, 199–208 (2009)
Van Valen, L. A new evolutionary law. Evol. Theory 1, 1–30 (1973)
Acknowledgements
We thank the Tucson Marine Phage Lab and B. Hurwitz for early access to the Pacific Ocean virome datasets and J. Yan for her assistance in isolating HTVC011P. This work was supported by an investigator award to S.J.G. from the Gordon and Betty Moore Foundation Marine Microbiology Initiative.
Author information
Authors and Affiliations
Contributions
Y.Z. isolated phages and performed genome sequencing and annotation; B.T. designed and implemented metagenomic bioinformatic analyses and prepared the manuscript; J.C.T. performed phylogenetic and metagenomic analyses; M.S.S. began the project and isolated the first virus, HTVC011P; Y.Z., M.E. and T.D. performed transmission electron microscopy; K.L.V. sequenced the HTVC011P genome; Z.C.L. assembled and annotated the viral genomes. M.B.S. and S.J.G. gave technical support and conceptual advice. S.J.G. assisted in writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-11, Supplementary Methods, Supplementary Tables 1-3 and Supplementary References. (PDF 6255 kb)
Rights and permissions
About this article
Cite this article
Zhao, Y., Temperton, B., Thrash, J. et al. Abundant SAR11 viruses in the ocean. Nature 494, 357–360 (2013). https://doi.org/10.1038/nature11921
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11921
This article is cited by
-
Mortality by ribosomal sequencing (MoRS) provides a window into taxon-specific cell lysis
The ISME Journal (2023)
-
Novel pelagiphage isolate Polarivirus skadi is a polar specialist that dominates SAR11-associated bacteriophage communities at high latitudes
The ISME Journal (2023)
-
Genetic engineering of marine cyanophages reveals integration but not lysogeny in T7-like cyanophages
The ISME Journal (2022)
-
Dynamics of actively dividing prokaryotes in the western Mediterranean Sea
Scientific Reports (2022)
-
Newly identified HMO-2011-type phages reveal genomic diversity and biogeographic distributions of this marine viral group
The ISME Journal (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.