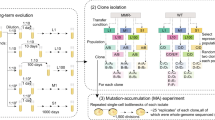
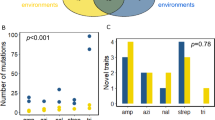
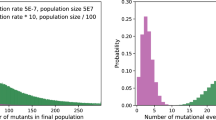
Abstract
The extinction rate of populations is predicted to rise under increasing rates of environmental change1,2,3. If a population experiencing increasingly stressful conditions lacks appropriate phenotypic plasticity or access to more suitable habitats, then genetic change may be the only way to avoid extinction1. Evolutionary rescue from extinction occurs when natural selection enriches a population for more stress-tolerant genetic variants1,3. Some experimental studies have shown that lower rates of environmental change lead to more adapted populations or fewer extinctions4,5,6,7,8,9. However, there has been little focus on the genetic changes that underlie evolutionary rescue. Here we demonstrate that some evolutionary trajectories are contingent on a lower rate of environmental change. We allowed hundreds of populations of Escherichia coli to evolve under variable rates of increase in concentration of the antibiotic rifampicin. We then genetically engineered all combinations of mutations from isolates evolved under lower rates of environmental change. By assessing fitness of these engineered strains across a range of drug concentrations, we show that certain genotypes are evolutionarily inaccessible under rapid environmental change. Rapidly deteriorating environments not only limit mutational opportunities by lowering population size, but they can also eliminate sets of mutations as evolutionary options. As anthropogenic activities are leading to environmental change at unprecedented rapidity1, it is critical to understand how the rate of environmental change affects both demographic and genetic underpinnings of evolutionary rescue.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bell, G. & Collins, S. Adaptation, extinction and global change. Evol. Appl. 1, 3–16 (2008)
Bellard, C., Bertelsmeier, C., Leadley, P., Thuiller, W. & Courchamp, F. Impacts of climate change on the future of biodiversity. Ecol. Lett. 15, 365–377 (2012)
Gomulkiewicz, R. & Holt, R. When does evolution by natural selection prevent extinction? Evolution 49, 201–207 (1995)
Bell, G. & Gonzalez, A. Evolutionary rescue can prevent extinction following environmental change. Ecol. Lett. 12, 942–948 (2009)
Bell, G. & Gonzalez, A. Adaptation and evolutionary rescue in metapopulations experiencing environmental deterioration. Science 332, 1327–1330 (2011)
Collins, S. & de Meaux, J. Adaptation to different rates of environmental change in Chlamydomonas. Evolution 63, 2952–2965 (2009)
Dallinger, W. H. The President’s Address. J. R. Microsc. Soc. 7, 185–199 (1887)
Perron, G. G., Gonzalez, A. & Buckling, A. The rate of environmental change drives adaptation to an antibiotic sink. J. Evol. Biol. 21, 1724–1731 (2008)
Huey, R., Partridge, L. & Fowler, K. Thermal sensitivity of Drosophila melanogaster responds rapidly to laboratory natural selection. Evolution 45, 751–756 (1991)
Gullberg, E. et al. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7, e1002158 (2011)
Reynolds, M. G. Compensatory evolution in rifampin-resistant Escherichia coli. Genetics 156, 1471–1481 (2000)
MacLean, R. C., Perron, G. G. & Gardner, A. Diminishing returns from beneficial mutations and pervasive epistasis shape the fitness landscape for rifampicin resistance in Pseudomonas aeruginosa. Genetics 186, 1345–1354 (2010)
Hall, A. R. & MacLean, R. C. Epistasis buffers the fitness effects of rifampicin resistance mutations in Pseudomonas aeruginosa. Evolution 65, 2370–2379 (2011)
Nei, M. & Li, W. H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl Acad. Sci. USA 76, 5269–5273 (1979)
Gillespie, J. H. A simple stochastic gene substitution model. Theor. Popul. Biol. 23, 202–215 (1983)
Weinreich, D. M., Watson, R. A. & Chao, L. Perspective: sign epistasis and genetic constraint on evolutionary trajectories. Evolution 59, 1165–1174 (2005)
Remold, S. K. & Lenski, R. E. Pervasive joint influence of epistasis and plasticity on mutational effects in Escherichia coli. Nature Genet. 36, 423–426 (2004)
Weinreich, D. M., Delaney, N. F., DePristo, M. A. & Hartl, D. L. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114 (2006)
Trindade, S. et al. Positive epistasis drives the acquisition of multidrug resistance. PLoS Genet. 5, e1000578 (2009)
Lalić, J. & Elena, S. F. Magnitude and sign epistasis among deleterious mutations in a positive-sense plant RNA virus. Heredity 109, 71–77 (2012)
Kvitek, D. J. & Sherlock, G. Reciprocal sign epistasis between frequently experimentally evolved adaptive mutations causes a rugged fitness landscape. PLoS Genet. 7, e1002056 (2011)
Lalić, J. & Elena, S. F. Epistasis between mutations is host-dependent for an RNA virus. Biol. Lett. 9, 20120396 (2013)
Comas, I. et al. Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nature Genet. 44, 106–110 (2012)
Martinez, J. L. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ. Pollut. 157, 2893–2902 (2009)
Gagneux, S. et al. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science 312, 1944–1946 (2006)
Sandgren, A. et al. Tuberculosis Drug Resistance Mutation Database. PLoS Med. 6, e1000002 (2009)
Bodbyl Roels, S. A. & Kelly, J. K. Rapid evolution caused by pollinator loss in Mimulus guttatus. Evolution 65, 2541–2552 (2011)
Skelly, D. K. et al. Evolutionary responses to climate change. Conserv. Biol. 21, 1353–1355 (2007)
Klironomos, J. N. et al. Abrupt rise in atmospheric CO2 overestimates community response in a model plant–soil system. Nature 433, 621–624 (2005)
Link, A. J., Phillips, D. & Church, G. M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179, 6228–6237 (1997)
Acknowledgements
We thank T. Bradshaw, J. Martiny and J. Tewksbury for sharing ideas that inspired this work; the Church laboratory for supplying the pKOV vector; C. Adams, S. DeCew, K. Dickinson, S. Drescher, C. Eshelman, K. Hobbs, C. Muerdter, B. Rogers and C. Shyue for help in the laboratory; and F. Bertels, C. Eshelman, C. Glenney and S. Singhal for comments on the manuscript. This material is based in part on work supported by the National Science Foundation under Cooperative Agreement Number DBI-0939454, a NSF CAREER Award Grant (DEB0952825), and a UW Royalty Research Fund Award (A74107).
Author information
Authors and Affiliations
Contributions
B.K. and S.T. designed the evolution experiment; S.T. and H.A.L. performed the evolution experiment; all authors designed and troubleshot the genetic and phenotypic protocols; H.A.L. performed the genetic work and phenotypic assays; B.K. did the mathematical and statistical analysis, and all authors contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary Results and Data, Supplementary Tables 1-2 and Supplementary Figures 1-3. (PDF 608 kb)
Rights and permissions
About this article
Cite this article
Lindsey, H., Gallie, J., Taylor, S. et al. Evolutionary rescue from extinction is contingent on a lower rate of environmental change. Nature 494, 463–467 (2013). https://doi.org/10.1038/nature11879
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11879
This article is cited by
-
Resistance mechanism of Escherichia coli strains with different ampicillin resistance levels
Applied Microbiology and Biotechnology (2024)
-
Environmental modulation of global epistasis in a drug resistance fitness landscape
Nature Communications (2023)
-
Population dynamics hide phenotypic changes driven by subtle chemical exposures: implications for risk assessments
Ecotoxicology (2023)
-
Hybridization drives genetic erosion in sympatric desert fishes of western North America
Heredity (2019)
-
Sign epistasis caused by hierarchy within signalling cascades
Nature Communications (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.