Abstract
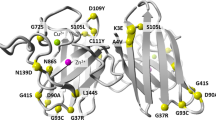
Amyotrophic lateral sclerosis (ALS) is a late-onset neurodegenerative disorder resulting from motor neuron death. Approximately 10% of cases are familial (FALS), typically with a dominant inheritance mode. Despite numerous advances in recent years1,2,3,4,5,6,7,8,9, nearly 50% of FALS cases have unknown genetic aetiology. Here we show that mutations within the profilin 1 (PFN1) gene can cause FALS. PFN1 is crucial for the conversion of monomeric (G)-actin to filamentous (F)-actin. Exome sequencing of two large ALS families showed different mutations within the PFN1 gene. Further sequence analysis identified 4 mutations in 7 out of 274 FALS cases. Cells expressing PFN1 mutants contain ubiquitinated, insoluble aggregates that in many cases contain the ALS-associated protein TDP-43. PFN1 mutants also display decreased bound actin levels and can inhibit axon outgrowth. Furthermore, primary motor neurons expressing mutant PFN1 display smaller growth cones with a reduced F/G-actin ratio. These observations further document that cytoskeletal pathway alterations contribute to ALS pathogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Kabashi, E. et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nature Genet. 40, 572–574 (2008)
Sreedharan, J. et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 (2008)
Kwiatkowski, T. J., Jr et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205–1208 (2009)
Vance, C. et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323, 1208–1211 (2009)
Johnson, J. O. et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 68, 857–864 (2010)
DeJesus-Hernandez, M. et al. Expanded GGGGCC exanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011)
Deng, H. X. et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477, 211–215 (2011)
Renton, A. E. et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011)
Gijselinck, I. et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 11, 54–65 (2012)
Mockrin, S. C. & Korn, E. D. Acanthamoeba profilin interacts with G-actin to increase the rate of exchange of actin-bound adenosine 5′-triphosphate. Biochemistry 19, 5359–5362 (1980)
Landers, J. E. et al. Reduced expression of the Kinesin-Associated Protein 3 (KIFAP3) gene increases survival in sporadic amyotrophic lateral sclerosis. Proc. Natl Acad. Sci. USA 106, 9004–9009 (2009)
Suetsugu, S. et al. The essential role of profilin in the assembly of actin for microspike formation. EMBO J. 17, 6516–6526 (1998)
Giesemann, T. et al. A role for polyproline motifs in the spinal muscular atrophy protein SMN. J. Biol. Chem. 274, 37908–37914 (1999)
Schutt, C. E. et al. The structure of crystalline profilin–β-actin. Nature 365, 810–816 (1993)
Wills, Z. et al. Profilin and the Abl tyrosine kinase are required for motor axon outgrowth in the Drosophila embryo. Neuron 22, 291–299 (1999)
Witke, W. et al. In mouse brain profilin I and profilin II associate with regulators of the endocytic pathway and actin assembly. EMBO J. 17, 967–976 (1998)
Braun, A. et al. Genomic organization of profilin-III and evidence for a transcript expressed exclusively in testis. Gene 283, 219–225 (2002)
Tilney, L. G. et al. Actin from Thyone sperm assembles on only one end of an actin filament: a behavior regulated by profilin. J. Cell Biol. 97, 112–124 (1983)
Takeuchi, H. et al. Hsp70 and Hsp40 improve neurite outgrowth and suppress intracytoplasmic aggregate formation in cultured neuronal cells expressing mutant SOD1. Brain Res. 949, 11–22 (2002)
Duan, W. et al. MG132 enhances neurite outgrowth in neurons overexpressing mutant TAR DNA-binding protein-43 via increase of HO-1. Brain Res. 1397, 1–9 (2011)
Fujii, R. et al. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr. Biol. 15, 587–593 (2005)
Witke, W. The role of profilin complexes in cell motility and other cellular processes. Trends Cell Biol. 14, 461–469 (2004)
Shao, J. et al. Phosphorylation of profilin by ROCK1 regulates polyglutamine aggregation. Mol. Cell. Biol. 28, 5196–5208 (2008)
Al-Chalabi, A. et al. Deletions of the heavy neurofilament subunit tail in amyotrophic lateral sclerosis. Hum. Mol. Genet. 8, 157–164 (1999)
Gros-Louis, F. et al. A frameshift deletion in peripherin gene associated with amyotrophic lateral sclerosis. J. Biol. Chem. 279, 45951–45956 (2004)
Puls, I. et al. Mutant dynactin in motor neuron disease. Nature Genet. 33, 455–456 (2003)
Hazan, J. et al. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nature Genet. 23, 296–303 (1999)
Reid, E. et al. A kinesin heavy chain (KIF5A) mutation in hereditary spastic paraplegia (SPG10). Am. J. Hum. Genet. 71, 1189–1194 (2002)
Mersiyanova, I. V. et al. A new variant of Charcot-Marie-Tooth disease type 2 is probably the result of a mutation in the neurofilament-light gene. Am. J. Hum. Genet. 67, 37–46 (2000)
Perrot, R. & Eyer, J. Neuronal intermediate filaments and neurodegenerative disorders. Brain Res. Bull. 80, 282–295 (2009)
Gudbjartsson, D. F. et al. Allegro, a new computer program for multipoint linkage analysis. Nature Genet. 25, 12–13 (2000)
Choi, M. et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl Acad. Sci. USA 106, 19096–19101 (2009)
Li, R. et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 (2009)
Li, R. et al. SNP detection for massively parallel whole-genome resequencing. Genome Res. 19, 1124–1132 (2009)
Kumar, P. et al. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols 4, 1073–1081 (2009)
The 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010)
Bosco, D. A. et al. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nature Neurosci. 13, 1396–1403 (2010)
Fallini, C. et al. High-efficiency transfection of cultured primary motor neurons to study protein localization, trafficking, and function. Mol. Neurodegener. 5, 17 (2010)
Meijering, E. et al. Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A 58A, 167–176 (2004)
Acknowledgements
Support was provided by the ALS Therapy Alliance, Project ALS, P2ALS, the Angel Fund, the Pierre L. de Bourgknecht ALS Research Foundation, the Al-Athel ALS Research Foundation, the ALS Family Charitable Foundation and a Francesco Caleffi donation (N.T. and V.S.). Grant support was provided by the National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke (NINDS) (1R01NS065847 (J.E.L.), 1R01NS050557 (R.H.B.), and RC2-NS070-342 (R.H.B.)), Muscular Dystrophy Association (MDA173851 (W.R.)) and AriSLA co-financed with support of ‘5x1000’—Healthcare research of the Ministry of Health (EXOMEFALS (N.T., C.G., V.S. and J.E.L.)). Support was provided by an SMA Europe fellowship to C.F. P.C.S. was supported through the auspices of H. R. Horvitz (Massachusetts Institute of Technology), an investigator of the Howard Hughes Medical Institute. We thank the laboratory of S. Doxsey, the UMass Medical School Digital Light Microscopy Core, the UMass Medical School Deep Sequencing Core, the Emory University Neuropathology Core, and M. Gearing and D. Cooper for their assistance.
Author information
Authors and Affiliations
Contributions
Sample collection, preparation and clinical evaluation: N.T., P.C.S., D.M.-Y., F.T., C.T., J.D.G., G.S., F.S., V.M., A.R., C.G., V.S., V.E.D., R.H.B. Performed experiments and data analysis: C.-H.W., C.F., N.T., P.J.K., P.C.S., K.P., P.L., D.M.B., J.E.K., P.G.-P., A.D.F., M.K., J.A., F.T., C.T., A.L.L., S.C.C., E.T.C., D.A.B., J.E.L. Scientific planning and direction: C.-H.W., C.F., N.T., D.M., M.J.M., J.A.Z., Z.-S.X., L.H.V, J.D.G., D.B.G., V.M., W.R., A.R., C.G., D.A.B., G.J.B., V.S., V.E.D., R.H.B., J.E.L. Initial manuscript preparation: C.-H.W., C.F., N.T., W.R., D.A.B., G.J.B., V.S., R.H.B., J.E.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-9 and Supplementary Figures 1-15. (PDF 16321 kb)
Rights and permissions
About this article
Cite this article
Wu, CH., Fallini, C., Ticozzi, N. et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature 488, 499–503 (2012). https://doi.org/10.1038/nature11280
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11280
This article is cited by
-
Physiological and pathological effects of phase separation in the central nervous system
Journal of Molecular Medicine (2024)
-
Amyotrophic lateral sclerosis: translating genetic discoveries into therapies
Nature Reviews Genetics (2023)
-
Genetics of amyotrophic lateral sclerosis: seeking therapeutic targets in the era of gene therapy
Journal of Human Genetics (2023)
-
Re-engineering the adenine deaminase TadA-8e for efficient and specific CRISPR-based cytosine base editing
Nature Biotechnology (2023)
-
Cellular Conversations in Glioblastoma Progression, Diagnosis and Treatment
Cellular and Molecular Neurobiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.