Abstract
Malaria elimination strategies require surveillance of the parasite population for genetic changes that demand a public health response, such as new forms of drug resistance1,2. Here we describe methods for the large-scale analysis of genetic variation in Plasmodium falciparum by deep sequencing of parasite DNA obtained from the blood of patients with malaria, either directly or after short-term culture. Analysis of 86,158 exonic single nucleotide polymorphisms that passed genotyping quality control in 227 samples from Africa, Asia and Oceania provides genome-wide estimates of allele frequency distribution, population structure and linkage disequilibrium. By comparing the genetic diversity of individual infections with that of the local parasite population, we derive a metric of within-host diversity that is related to the level of inbreeding in the population. An open-access web application has been established for the exploration of regional differences in allele frequency and of highly differentiated loci in the P. falciparum genome.
Similar content being viewed by others
Main
The genetic diversity and evolutionary plasticity of P. falciparum are major obstacles for malaria elimination. New forms of resistance against antimalarial drugs are continually emerging1,2, and new forms of antigenic variation are a critical point of vulnerability for future malaria vaccines. Effective tools are needed to detect evolutionary changes in the parasite population and to monitor the spread of genetic variants that affect malaria control.
Here we describe the use of deep sequencing to analyse P. falciparum diversity, using blood samples from patients with malaria. The P. falciparum genome has several unusual features that greatly complicate sequence analysis, such as extreme AT bias, large tracts of non-unique sequence and several large families of intensely polymorphic genes3. Our aim was therefore not to determine the entire genome sequence of individual field samples—which would be prohibitively expensive with current technologies—but to define an initial set of single nucleotide polymorphisms (SNPs) distributed across the P. falciparum genome, whose genotype can be ascertained with confidence in parasitized blood samples by deep sequencing.
An additional complication in the analysis of P. falciparum genome variation is that the billions of haploid parasites that infect a single individual can be a complex mixture of genetic types. Previous studies4,5,6,7,8 have largely focused on laboratory-adapted parasite clones, but the within-host diversity of natural infections is of fundamental biological interest. Parasites in the blood replicate asexually, but when they are taken up in the blood meal of an Anopheles mosquito they undergo sexual mating. If the parasites in the blood are of diverse genetic types, this process of sexual mating can generate novel recombinant forms. Deep sequencing provides new ways of investigating within-host diversity and the role of sexual recombination in parasite evolution.
P. falciparum DNA was obtained from blood samples collected from 290 patients with malaria at clinics in Burkina Faso, Cambodia, Kenya, Mali, Papua New Guinea and Thailand (Supplementary Table 1). For 149 samples we used the conventional method of growing the parasites in short-term blood culture before extracting the P. falciparum DNA. For 141 samples we used a new method by which P. falciparum DNA is extracted directly from venous blood samples after the removal of leukocytes9. We refer to these as cultured and direct samples, respectively.
Paired-end sequence reads were generated (median 7 × 108 base pairs per sample) by using the Illumina Genome Analyzer platform. Sequence analysis was divided into stages of SNP discovery, quality control filtering, genotyping and validation (see Supplementary Methods and Supplementary Fig. 1). After alignment to the 3D7 reference genome3, non-coding regions had a much lower read depth than coding regions (Supplementary Fig. 2): this can be ascribed to their high AT content (non-coding 87% AT, coding 70% AT). Read depth was also low in the highly polymorphic var, rifin and stevor coding regions (Supplementary Fig. 3). For the purposes of this study, to decrease genotyping errors due to low coverage or copy number variation we excluded all non-coding regions, as well as coding regions at the extremes of the read depth distribution. After these exclusions we were left with 70% of all exonic positions across the genome, with more than 50% of exonic positions for 71% of genes, and more than 70% for 54% of genes (Supplementary Table 2).
Within-host diversity complicates the process of excluding sequencing and alignment errors that are manifested as false heterozygous genotypes. Two approaches were identified to address this problem (see Supplementary Methods). We scored each position in the reference genome for its degree of uniqueness, and this was found to be a strong predictor of false heterozygous genotypes. We also observed a relationship between the population allele frequency of a SNP and its average level of within-sample heterozygosity, analogous to the Hardy–Weinberg relationship in diploid organisms. This enabled us to exclude SNPs that had excessive levels of within-sample heterozygosity relative to their population frequency.
After applying the above filters, and excluding SNPs and samples with high levels of missing data, we obtained a final data set of 86,158 SNPs genotyped in 227 samples (120 direct and 107 cultured) in which a median of 98% samples had valid genotyping data for each SNP, and a median of 98% SNPs had valid genotyping data for each sample (Supplementary Fig. 4). This set of 86,158 SNPs (here referred to as the 86k SNP set) represents 10% of the SNPs discovered at the initial stage of sequence alignment. Comparison with the PlasmoDB 5.5 database indicates that 77,283 (89%) of these SNPs are novel, but it should be noted that previous genome-wide SNP discovery efforts have largely been based on low-coverage capillary sequencing, and the overall error rate is unknown4,5,6.
The accuracy of genotype calls in the 86k SNP set was evaluated by five independent approaches (see Supplementary Methods). We examined the evidence for 275 putative novel SNPs using independent data from PCR-based capillary sequencing and Sequenom primer-extension mass spectrometry: the existence of the novel allele was confirmed for 270 of the 275 loci. The genotype concordance rate with Sequenom was 99.9% and with capillary sequencing it was 98.6%, excluding heterozygotes (Supplementary Tables 3 and 4). In the case of heterozygous genotypes, deep sequencing gives the allelic ratio, whereas most other P. falciparum SNP typing methods give the majority allele or return a missing genotype. The observation of heterozygosity by deep sequencing was correlated with Sequenom’s failing to call a majority allele, but when Sequenom made a majority allele call it agreed with deep sequencing data in 94.8% of cases (Supplementary Fig. 5). Capillary sequencing data do not allow allelic ratios to be quantified precisely, but visual inspection of capillary sequence traces was consistent with heterozygous genotype calls in the deep sequencing data (Supplementary Fig. 6). In a separate study to be reported elsewhere, we sequenced 90 laboratory-adapted parasite clones derived from three genetic crosses of P. falciparum and determined that the rate of Mendelian errors in the 86k SNP set was 0.05%.
Population genetic analyses were conducted with the 86k SNP set typed in 227 samples as described above. The allele frequency spectrum was dominated by low-frequency variants (Fig. 1 and Supplementary Fig. 7) even when synonymous sites alone were considered, which is consistent with recent population expansion (Supplementary Table 5)10. Samples from Africa had a greater number of low-frequency variants than samples from Southeast Asia or Papua New Guinea with or without correction for sample size. Multiple lines of evidence indicate that P. falciparum originated in Africa, and loss of low-frequency variation might have occurred as a result of population bottlenecks during migration out of Africa, as in human populations10,11.
a, Minor-allele frequency distribution of 86k SNPs set in samples from different continents: Africa (red), Southeast Asia (green) and Papua New Guinea (blue). The y axis shows the number of SNPs in each category of allele frequency. Supplementary Figure 7 shows the data corrected for sample size. b, Ratio of non-synonymous (N) to synonymous (S) substitutions, as a function of derived allele frequency for SNPs that are private to either Africa, Southeast Asia or Papua New Guinea.
The most likely ancestral state of each SNP was determined from the P. reichenowi genome sequence but is difficult to estimate with confidence, because P. reichenowi might have diverged from P. falciparum relatively recently, and its genome sequence has been determined for only one individual (refs 6, 12 and T. D. Otto, unpublished obervations). There seem to be more SNPs with low-frequency derived (non-ancestral) alleles in Africa than in Southeast Asia or Papua New Guinea (Supplementary Figs 8 and 9). Focusing on SNPs that are private to one continent, those with high derived allele frequency show a considerable excess of non-synonymous substitutions, suggesting that these are largely the result of directional selection (Fig. 1b and Supplementary Fig. 10).
Many SNPs (64%) were observed in only one continent, but most were low-frequency variants and larger sample sizes are needed to determine how many of these are truly private. Corrected for sample size, the number of private SNPs was greatest in East Africa and least in Southeast Asia, both of which comprised cultured samples (Supplementary Fig. 11). Intermediate numbers were observed in West Africa and Papua New Guinea, both of which comprised direct samples. Thus the effect of culturing on SNP ascertainment seems to be relatively small in comparison with the effect of geographical location.
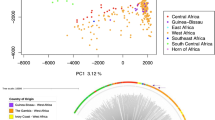
The global population structure of P. falciparum shows a clear division by continent (Fig. 2a). Mean fixation index (Fst) values between continents ranged from 0.19 to 0.28 (Supplementary Table 6). Population structure within continents is evident from Fst values, principal-components analysis (Supplementary Fig. 12) and a neighbour-joining tree (Fig. 2b). All of these methods show greater degree of population structure in Southeast Asia than in West Africa; that is, samples from Cambodia and Thailand form separate clusters, whereas samples from Mali and Burkina Faso are intermixed. These data are consistent with previous evidence that parasite population structure tends to be increased in regions of low or patchy malaria transmission13.
a, Principal-components analysis. b, Unrooted neighbour-joining tree. Leaf branches are coloured (as in a) according to the country of origin of the sample.
To understand the hierarchical population structure of P. falciparum, methods are needed to quantify the genetic diversity of individual infections relative to the genetic diversity of the parasite population as a whole. With deep sequencing data, we can estimate levels of heterozygosity both within an individual sample (Hw) and within the local parasite population (Hs). For a biallelic SNP, we define Hw as 2pwqw, where pw and qw denote the proportions of the two alleles in the sequence reads of an individual sample, and Hs as 2psqs, where ps and qs denote the corresponding population allele frequencies at that geographical location. We observe a strong linear relationship between Hw and Hs when data for all 86k SNPs are aggregated for an individual sample (Fig. 3a and Supplementary Fig. 13). More specifically, each sample shows a linear relationship between Hw and Hs but the gradient of the line varies considerably between samples. This gradient is essentially a genome-wide estimate of Hw/Hs for the sample in question. Thus for each sample we can derive the metric Fws, where

This is closely related to Wright’s inbreeding coefficient Fis, which can be formulated as

where Hi is the heterozygosity of the individual and Hs is that of the local population14. Estimation of Fis is of practical relevance for malaria control, because high rates of inbreeding are thought to favour the emergence of multigenic drug resistance15,16. Fis is conventionally measured at the oocyst stage of infection—that is, after the parasites have undergone sexual mating within the mosquito and before they develop into separate haploid forms—but this is technically demanding and difficult to implement on a large scale15,17. Because parasites undergo sexual mating shortly after the mosquito has ingested blood from an infected person, the level of within-host diversity determines the potential for inbreeding or outcrossing in the next generation. Thus Fws values observed in blood samples provide a proxy indicator of inbreeding rates in the population. The precise relationship to inbreeding rates quantified in oocysts merits further investigation. We report elsewhere a study of how Fws relates to standard methods of estimating multiplicity of infection18.
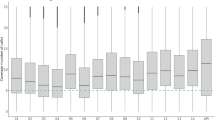
a, Relationship between within-host heterozygosity (Hw) and heterozygosity in the local parasite population (Hs) for all samples in the West African population. Each line represents a different sample, whose within-host heterozygosity values were averages across all SNPs, categorized according to their heterozygosity in the local parasite population. Separate plots for each population are shown in Supplementary Fig. 13. b, Box plot showing the distribution of Fws estimates in samples from each of the four populations.
We observe marked differences in Fws between locations (Fig. 3b). High levels of Fws (0.95 or more) were much more common in Papua New Guinea (89% of samples) than in West Africa (38%), with intermediate rates in Southeast Asia (67%) and East Africa (63%). Culturing might affect Fws estimation, but the samples from Papua New Guinea and West Africa were not cultured. In general, high levels of inbreeding tend to be associated with low transmission intensity13, and these data are therefore somewhat surprising because the entomological inoculation rate has been estimated to lie in the range 45–293 in Madang in Papua New Guinea19, where the Papua New Guinea samples were collected, in contrast with 140–389 in Burkina Faso19, about 6 in rural areas of Cambodia20 and about 1 on the Thailand–Burma border21. Although the entomological inoculation rate can be highly variable within a locality and these estimates are indicative, it seems unlikely that the high levels of Fws in Papua New Guinea are primarily due to low transmission intensity. An alternative explanation is that, in this geographical region, people tend to live in small isolated communities, which might reduce the likelihood of infection with parasites of different genetic types. The small size of the Papua New Guinea sample provides limited information about local parasite population structure (Supplementary Fig. 14), but previous studies indicate that this is very high in some villages within this area of Papua New Guinea22.
These data allow linkage disequilibrium in the P. falciparum genome to be estimated with greater precision than has previously been possible. In particular, we can begin to distinguish linkage disequilibrium due to haplotype structure, which decays with distance in the genome, from linkage disequilibrium due to population structure, which is independent of distance in the genome (see Supplementary Methods, Supplementary Tables 7 and 8 and Supplementary Figs 15–17). Averaged across the genome, after correcting for population structure and other confounders, we find that r2 decays to less than 0.1 within 1 kilobase (kb) in all populations studied here, whereas D′ decays to less than 0.1 within about 1 kb in West Africa and East Africa, and within 50 kb in Southeast Asia and Papua New Guinea (Supplementary Fig. 18). These findings imply that high levels of haplotypic diversity exist at all of these locations, despite low transmission intensity and high rates of inbreeding at some locations. This might be partly due to the high rate of meiotic recombination in P. falciparum, previously estimated to be about 17 kb per centimorgan23. It is also possible that much of the haplotypic diversity seen in contemporary P. falciparum populations has ancient origins, and arose in Africa before P. falciparum was spread around the world by human migration. This would be analogous to the situation that is seen in human populations, in which migration out of Africa was associated with a series of population bottlenecks, which have led to a reduction in haplotypic diversity in descendant populations around the world11. The higher levels of linkage disequilibrium observed in Southeast Asia and Papua New Guinea than in West Africa and East Africa are consistent with both of these possibilities.
A web application is provided for browsing, querying and downloading information about all of the SNPs genotyped in this study and their allele frequencies in different geographical regions (http://www.malariagen.net/resource/10). It can be used, for example, to view regional patterns of variation in known antimalarial drug resistance genes: from these data it is immediately apparent that the pfcrt K76T allele has markedly different haplotypic backgrounds in Southeast Asia and in Papua New Guinea, consistent with previous evidence that chloroquine resistance has evolved independently in multiple locations (Supplementary Table 9)1,24. It can also be used to search for genes that are highly differentiated between geographical regions (Supplementary Tables 10 and 11). For example, two genes that affect the fertility of gametocytes, Pfs230 and Pf47, are among the most highly differentiated loci in this data set25. Two SNPs in Pfs230 codon 1566 result in three amino-acid variants: N (widespread), T (private to Southeast Asia, frequency 0.87) and K (private to Africa, frequency 0.79). Codon variant T236I of Pf47 has a fixed difference between Africa and other populations. These data lend weight to previous reports of extreme differentiation in Pf47 and the related gene Pfs48/45 (ref. 26), which is suggested to be due to evolutionary selection of gamete recognition and compatibility. Another example is codon variant F368S of the putative transporter gene PFA0245w (ref. 27), which has a fixed difference between Papua New Guinea and other populations, raising the question of whether this has a function in drug resistance; it is also noteworthy that the Plasmodium berghei orthologue of this gene is critical for sexual development of the parasite28.
These data are the first stage in the development of methods for population-based genome sequencing of P. falciparum. Work is ongoing to increase the number of SNPs that can be reliably genotyped, and to develop accurate methods for typing indels, copy number polymorphisms and large structural variations. Future studies will benefit from new methods to reduce the effects of AT bias on sequencing library preparation29,30, and the increasing length and accuracy of sequencing reads will allow greater access to highly polymorphic regions of the genome. Such technical advances will enable an expanding range of applications, for example high-resolution analyses of local population structure to explore models of space–time clustering and immunological strain selection.
Genome sequencing of parasites in clinical blood samples is an important step towards translation to public health applications, for example developing effective genetic markers to track the spread of antimalarial drug resistance and to monitor evolutionary changes in the parasite population7,8. There is a need to develop protocols, tools and resources and to enable researchers in malaria endemic countries to integrate parasite genome sequencing into clinical and epidemiological investigations, and to facilitate open-access sharing of large-scale population genomic data.
Methods Summary
Blood samples from malaria patients were collected with informed consent after approval by local ethics committees. Parasite DNA was extracted from blood samples after leukocyte depletion to minimize contamination with human DNA, or after short-term culture in vitro. Samples with less than 60% human DNA contamination were sequenced with an Illumina Genome Analyser. Sequence reads of length 37–76 base pairs were aligned to the 3D7 reference sequence3 using the bwa and samtools algorithms, and then with the more stringent SNP-o-matic algorithm that allowed for SNPs discovered in the first step. This gave 868,117 potential SNPs, including 74% (71,608/96,527) of SNPs previously identified in the PlasmoDB 5.5 database.
Various quality-control steps were applied. We discarded potential SNPs with insufficient evidence, those in non-coding regions, and those in coding regions with sequencing coverage outside the 15th centile and the 85th centile of read depth. To minimize alignment errors, we scored each position in the reference genome for its degree of uniqueness, and excluded positions that were liable to give false heterozygous genotypes. We analysed levels of heterozygosity across all samples, discarding positions where heterozygosity was inconsistent with population allele frequencies. Genotypes were determined at positions with at least five reads, resulting in a set of 86,158 biallelic SNPs that could be genotyped with low missingness in 227 samples.
Five methods were used to validate genotyping calls: Sequenom primer-extension mass spectrometry, PCR-based capillary sequencing, Illumina GoldenGate array, high-density NimbleGen microarray, and analysis of error rates in genotypes from P. falciparum genetic crosses. Allele frequencies were determined in four populations, deriving ancestral alleles from comparison with P. reichenowi sequences wherever possible. SNPs were classified in accordance with PlasmoDB 5.5 functional annotations. Principal-components analysis and phylogeny analysis were performed using R language libraries, and custom R and Java programs were used for other data analysis.
Accession codes
Data deposits
All sequence data are available online at the European Nucleotide Archive (ENA); accession numbers are listed in Supplementary Table 12. An online catalogue of SNPs and allele frequencies is available at http://www.malariagen.net/resource/10.
References
Wootton, J. C. et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature 418, 320–323 (2002)
Dondorp, A. M. et al. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 361, 455–467 (2009)
Gardner, M. J. et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511 (2002)
Mu, J. et al. Genome-wide variation and identification of vaccine targets in the Plasmodium falciparum genome. Nature Genet. 39, 126–130 (2007)
Volkman, S. K. et al. A genome-wide map of diversity in Plasmodium falciparum. Nature Genet. 39, 113–119 (2007)
Jeffares, D. C. et al. Genome variation and evolution of the malaria parasite Plasmodium falciparum. Nature Genet. 39, 120–125 (2007)
Neafsey, D. E. et al. Genome-wide SNP genotyping highlights the role of natural selection in Plasmodium falciparum population divergence. Genome Biol. 9, R171 (2008)
Mu, J. et al. Plasmodium falciparum genome-wide scans for positive selection, recombination hot spots and resistance to antimalarial drugs. Nature Genet. 42, 268–271 (2010)
Auburn, S. et al. An effective method to purify Plasmodium falciparum DNA directly from clinical blood samples for whole genome high-throughput sequencing. PLoS ONE 6, e22213 (2011)
Joy, D. A. et al. Early origin and recent expansion of Plasmodium falciparum. Science 300, 318–321 (2003)
Li, J. Z. et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319, 1100–1104 (2008)
Prugnolle, F. et al. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc. Natl Acad. Sci. USA 107, 1458–1463 (2010)
Anderson, T. J. et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol. Biol. Evol. 17, 1467–1482 (2000)
Hartl, D. & Clark, A. G. Principles of population genetics 4th edn (Sinauer, 2007)
Paul, R. E. et al. Mating patterns in malaria parasite populations of Papua New Guinea. Science 269, 1709–1711 (1995)
Dye, C. & Williams, B. G. Multigenic drug resistance among inbred malaria parasites. Proc. R. Soc. Lond. B 264, 61–67 (1997)
Hill, W. G., Babiker, H. A., Ranford-Cartwright, L. C. & Walliker, D. Estimation of inbreeding coefficients from genotypic data on multiple alleles, and application to estimation of clonality in malaria parasites. Genet. Res. 65, 53–61 (1995)
Auburn, S. et al. Characterization of within-host Plasmodium falciparum diversity using next-generation sequence data. PLoS ONE 7, e32891 (2012)
Smith, D. L., Drakeley, C. J., Chiyaka, C. & Hay, S. I. A quantitative analysis of transmission efficiency versus intensity for malaria. Nature Commun. 1, 108 (2010)
Trung, H. D. et al. Malaria transmission and major malaria vectors in different geographical areas of Southeast Asia. Trop. Med. Int. Health 9, 230–237 (2004)
Paul, R. E. et al. Genetic analysis of Plasmodium falciparum infections on the north-western border of Thailand. Trans. R. Soc. Trop. Med. Hyg. 93, 587–593 (1999)
Schultz, L. et al. Multilocus haplotypes reveal variable levels of diversity and population structure of Plasmodium falciparum in Papua New Guinea, a region of intense perennial transmission. Malar. J. 9, 336 (2010)
Su, X. et al. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science 286, 1351–1353 (1999)
Mehlotra, R. K. et al. Evolution of a unique Plasmodium falciparum chloroquine-resistance phenotype in association with pfcrt polymorphism in Papua New Guinea and South America. Proc. Natl Acad. Sci. USA 98, 12689–12694 (2001)
van Dijk, M. R. et al. Three members of the 6-cys protein family of Plasmodium play a role in gamete fertility. PLoS Pathog. 6, e1000853 (2010)
Anthony, T. G., Polley, S. D., Vogler, A. P. & Conway, D. J. Evidence of non-neutral polymorphism in Plasmodium falciparum gamete surface protein genes Pfs47 and Pfs48/45. Mol. Biochem. Parasitol. 156, 117–123 (2007)
Martin, R. E., Henry, R. I., Abbey, J. L., Clements, J. D. & Kirk, K. The ‘permeome’ of the malaria parasite: an overview of the membrane transport proteins of Plasmodium falciparum. Genome Biol. 6, R26 (2005)
Boisson, B. et al. The novel putative transporter NPT1 plays a critical role in early stages of Plasmodium berghei sexual development. Mol. Microbiol. 81, 1343–1357 (2011)
Kozarewa, I. et al. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nature Methods 6, 291–295 (2009)
Oyola, S. O. et al. Optimizing Illumina Next-Generation Sequencing library preparation for extremely AT-biased genomes. BMC Genomics 13, 1 (2012)
Acknowledgements
We thank G. Dougan and N. Day for support, and T. Anderson and M. Mackinnon for comments. The sequencing and analysis components of this study were supported by the Wellcome Trust through Sanger Institute core funding (077012/Z/05/Z; 098051) and a Strategic Award (090770/Z/09/Z); the Medical Research Council (MRC) through the MRC Centre for Genomics and Global Health (G0600718) and an MRC Professorship to D.P.K. (G19/9). Other parts of this study were partly supported by the Wellcome Trust including core support to the Wellcome Trust Centre for Human Genetics (075491/Z/04; 090532/Z/09/Z); the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health; and a Howard Hughes Medical Institute International Scholarship (55005502) to A.D.
Author information
Authors and Affiliations
Contributions
S.A., S.C., A.D., O.D., I.Z., J.-B.O., P.M., I.M., P.S., A.N., S.B., S.M.K., K.M., H.J., X.-Z.S., C.A., R.F., D.S., F.N., M.I., N.J.W., L.A.-E., C.S., V.M., D.M., A.A.-N. and D.J.C. performed field and laboratory studies to obtain P. falciparum samples for sequencing. S.A., S.C., M.S., E.A., D.A., E.D., S.O., M.A.Q., D.J.T., B.M., C.I.N. and M.B. developed and implemented methods for sample processing and sequencing library preparation. J.A.-G., M.M., O.M., G.M., V.R.R. and D.J. developed software for data management and visualization. K.A.R., C.H., A.J., K.R., J.C.T., M.T.F., S.C., S.A., D.A., C.I.N. and M.B. performed validation experiments. C.V.P., S.T.-H. and C.R. contributed to development of the project. B.M., M.B., C.I.N. and J.C.R. provided project management and oversight. O.M., M.M., D.P.K., J.O.’B. and T.G.C. conducted data analyses. D.P.K. and O.M. developed the Fws metric. D.P.K., O.M. and M.M. wrote the manuscript and collated comments from all authors. S.A. and S.C. made equal contributions.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Methods, which include references, tables 1-5 and figures 1-9, Supplementary Tables 1-12 and Supplementary Figures 1-18 (see page 1 for details). (PDF 4937 kb)
Rights and permissions
About this article
Cite this article
Manske, M., Miotto, O., Campino, S. et al. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 487, 375–379 (2012). https://doi.org/10.1038/nature11174
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11174
This article is cited by
-
An optimized GATK4 pipeline for Plasmodium falciparum whole genome sequencing variant calling and analysis
Malaria Journal (2023)
-
Genomics of Plasmodium vivax in Colombia reveals evidence of local bottle-necking and inter-country connectivity in the Americas
Scientific Reports (2023)
-
Population dynamics and drug resistance mutations in Plasmodium falciparum on the Bijagós Archipelago, Guinea-Bissau
Scientific Reports (2023)
-
Analysis of sequence diversity in Plasmodium falciparum glutamic acid-rich protein (PfGARP), an asexual blood stage vaccine candidate
Scientific Reports (2023)
-
Unveiling the genomic landscape of malaria in natural infections
Nature Reviews Microbiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.