Abstract
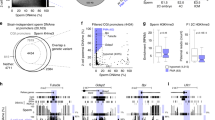
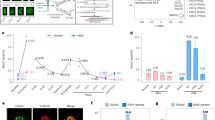
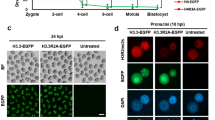
The modification of DNA by 5-methylcytosine (5mC) has essential roles in cell differentiation and development through epigenetic gene regulation1. 5mC can be converted to another modified base, 5-hydroxymethylcytosine (5hmC), by the tet methylcytosine dioxygenase (Tet) family of enzymes2,3. Notably, the balance between 5hmC and 5mC in the genome is linked with cell-differentiation processes such as pluripotency and lineage commitment4,5,6,7. We have previously reported that the maternal factor PGC7 (also known as Dppa3, Stella) is required for the maintenance of DNA methylation in early embryogenesis, and protects 5mC from conversion to 5hmC in the maternal genome8,9. Here we show that PGC7 protects 5mC from Tet3-mediated conversion to 5hmC by binding to maternal chromatin containing dimethylated histone H3 lysine 9 (H3K9me2) in mice. In addition, imprinted loci that are marked with H3K9me2 in mature sperm are protected by PGC7 binding in early embryogenesis. This type of regulatory mechanism could be involved in DNA modifications in somatic cells as well as in early embryos.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Feng, S., Jacobsen, S. E. & Reik, W. Epigenetic reprogramming in plant and animal development. Science 330, 622–627 (2010)
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009)
Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324, 929–930 (2009)
Koh, K. et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 8, 200–213 (2011)
Ficz, G. et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402 (2011)
Williams, K. et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473, 343–348 (2011)
Wu, H. et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473, 389–393 (2011)
Nakamura, T. et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nature Cell Biol. 9, 64–71 (2007)
Wossidlo, M. et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nature Commun. 2, 241 (2011)
Santos, F., Peters, A. H., Otte, A. P., Reik, W. & Dean, W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev. Biol. 280, 225–236 (2005)
Tachibana, M. et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16, 1779–1791 (2002)
Tsumura, A. et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11, 805–814 (2006)
Ge, Y. Z. et al. Chromatin targeting of de novo DNA methyltransferases by the PWWP domain. J. Biol. Chem. 279, 25447–25454 (2004)
Hajkova, P. et al. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329, 78–82 (2010)
Yamane, K. et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125, 483–495 (2006)
Olek, A. & Walter, J. The pre-implantation ontogeny of the H19 methylation imprint. Nature Genet. 17, 275–276 (1997)
Hammoud, S. S. et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460, 473–478 (2009)
Brykczynska, U. et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nature Struct. Mol. Biol. 17, 679–687 (2010)
Farthing, C. R. et al. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 4, e1000116 (2008)
Iqbal, K., Jin, S. G., Pfeifer, G. P. & Szabo, P. E. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl Acad. Sci. USA 108, 3642–3647 (2011)
Gu, T. et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477, 606–610 (2011)
Liu, Z. et al. Jmjd1a demethylase-regulated histone modification is essential for cAMP-response element modulator-refulated gene expression and spermatogenesis. J. Biol. Chem. 285, 2758–2770 (2010)
Ito, S. et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 (2011)
He, Y. F. et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 (2011)
Pogribny, I., Yi, P. & James, S. A sensitive new method for rapid detection of abnormal methylation patterns in global DNA and within CpG islands. Biochem. Biophys. Res. Commun. 262, 624–628 (1999)
Acknowledgements
We thank M. Okano, H. Niwa and H. Kimura for providing Dnmt1−/− Dnmt3a−/− Dnmt3b−/−ES cells, plasmids and antibody. We also thank N. Asada for assistance, and A. Mizokami and M. Imaizumi for secretarial assistance. This work was supported in part by grants from the Ministry of Education, Science, Sports, Culture and Technology of Japan.
Author information
Authors and Affiliations
Contributions
T. Nakamura and T. Nakano conceived the project and wrote the manuscript. T. Nakamura designed and performed the all experiments and evaluated the results. Y.-J.L., H.N. and H.U. propagated PGC7−/− mice. K.I., S.M. and A.O. performed some of the experiments. M.T. and Y.S. provided G9a−/− ES cells and related materials.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion, Supplementary References, Supplementary Figures 1-22 and Supplementary Tables 1-2. (PDF 7551 kb)
Rights and permissions
About this article
Cite this article
Nakamura, T., Liu, YJ., Nakashima, H. et al. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature 486, 415–419 (2012). https://doi.org/10.1038/nature11093
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11093
This article is cited by
-
Interaction network of human early embryonic transcription factors
EMBO Reports (2024)
-
Auto-suppression of Tet dioxygenases protects the mouse oocyte genome from oxidative demethylation
Nature Structural & Molecular Biology (2024)
-
Dynamics of DNA hydroxymethylation and methylation during mouse embryonic and germline development
Nature Genetics (2023)
-
Dppa3 Improves the Germline Competence of Pluripotent Stem Cells
Stem Cell Reviews and Reports (2023)
-
Regulation of paternal 5mC oxidation and H3K9me2 asymmetry by ERK1/2 in mouse zygotes
Cell & Bioscience (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.