Abstract
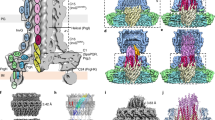
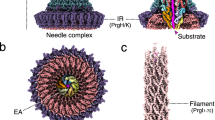
Pathogenic bacteria using a type III secretion system (T3SS)1,2 to manipulate host cells cause many different infections including Shigella dysentery, typhoid fever, enterohaemorrhagic colitis and bubonic plague. An essential part of the T3SS is a hollow needle-like protein filament through which effector proteins are injected into eukaryotic host cells3,4,5,6. Currently, the three-dimensional structure of the needle is unknown because it is not amenable to X-ray crystallography and solution NMR, as a result of its inherent non-crystallinity and insolubility. Cryo-electron microscopy combined with crystal or solution NMR subunit structures has recently provided a powerful hybrid approach for studying supramolecular assemblies7,8,9,10,11,12, resulting in low-resolution and medium-resolution models13,14,15,16,17. However, such approaches cannot deliver atomic details, especially of the crucial subunit–subunit interfaces, because of the limited cryo-electron microscopic resolution obtained in these studies. Here we report an alternative approach combining recombinant wild-type needle production, solid-state NMR, electron microscopy and Rosetta modelling to reveal the supramolecular interfaces and ultimately the complete atomic structure of the Salmonella typhimurium T3SS needle. We show that the 80-residue subunits form a right-handed helical assembly with roughly 11 subunits per two turns, similar to that of the flagellar filament of S. typhimurium. In contrast to established models of the needle in which the amino terminus of the protein subunit was assumed to be α-helical and positioned inside the needle, our model reveals an extended amino-terminal domain that is positioned on the surface of the needle, while the highly conserved carboxy terminus points towards the lumen.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Galan, J. E. & Wolf-Watz, H. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444, 567–573 (2006)
Cornelis, G. R. The type III secretion injectisome. Nature Rev. Microbiol. 4, 811–825 (2006)
Kubori, T. et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280, 602–605 (1998)
Kimbrough, T. G. & Miller, S. I. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl Acad. Sci. USA 97, 11008–11013 (2000)
Tamano, K. et al. Supramolecular structure of the Shigella type III secretion machinery: the needle part is changeable in length and essential for delivery of effectors. EMBO J. 19, 3876–3887 (2000)
Blocker, A. J. et al. What’s the point of the type III secretion system needle? Proc. Natl Acad. Sci. USA 105, 6507–6513 (2008)
Nogales, E. & Grigorieff, N. Molecular machines: putting the pieces together. J. Cell Biol. 152, F1–F10 (2001)
Volkmann, N. & Hanein, D. Docking of atomic models into reconstructions from electron microscopy. Methods Enzymol. 374, 204–225 (2003)
Rossmann, M. G., Morais, M. C., Leiman, P. G. & Zhang, W. Combining X-ray crystallography and electron microscopy. Structure 13, 355–362 (2005)
Spreter, T. et al. A conserved structural motif mediates formation of the periplasmic rings in the type III secretion system. Nature Struct. Mol. Biol. 16, 468–476 (2009)
Baker, M. L., Zhang, J., Ludtke, S. J. & Chiu, W. Cryo-EM of macromolecular assemblies at near-atomic resolution. Nature Protocols 5, 1697–1708 (2010)
Schraidt, O. & Marlovits, T. C. Three-dimensional model of Salmonella’s needle complex at subnanometer resolution. Science 331, 1192–1195 (2011)
Fujii, T., Iwane, A. H., Yanagida, T. & Namba, K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature 467, 724–728 (2010)
Yonekura, K., Maki-Yonekura, S. & Namba, K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424, 643–650 (2003)
Wang, H. W. & Nogales, E. Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Nature 435, 911–915 (2005)
Craig, L. et al. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol. Cell 23, 651–662 (2006)
Deane, J. E. et al. Molecular model of a type III secretion system needle: implications for host-cell sensing. Proc. Natl Acad. Sci. USA 103, 12529–12533 (2006)
Poyraz, O. et al. Protein refolding is required for assembly of the type three secretion needle. Nature Struct. Mol. Biol. 17, 788–792 (2010)
Loquet, A., Lv, G., Giller, K., Becker, S. & Lange, A. 13C spin dilution for simplified and complete solid-state NMR resonance assignment of insoluble biological assemblies. J. Am. Chem. Soc. 133, 4722–4725 (2011)
Hong, M. Determination of multiple φ-torsion angles in proteins by selective and extensive 13C labeling and two-dimensional solid-state NMR. J. Magn. Reson. 139, 389–401 (1999)
Lundstrom, P. et al. Fractional 13C enrichment of isolated carbons using [1-13C]- or [2-13C]-glucose facilitates the accurate measurement of dynamics at backbone Cα and side-chain methyl positions in proteins. J. Biomol. NMR 38, 199–212 (2007)
Lewandowski, J. R., De Paepe, G. & Griffin, R. G. Proton assisted insensitive nuclei cross polarization. J. Am. Chem. Soc. 129, 728–729 (2007)
Wasmer, C. et al. Amyloid fibrils of the HET-s(218–289) prion form a beta solenoid with a triangular hydrophobic core. Science 319, 1523–1526 (2008)
Loquet, A., Giller, K., Becker, S. & Lange, A. Supramolecular interactions probed by 13C–13C solid-state NMR spectroscopy. J. Am. Chem. Soc. 132, 15164–15166 (2010)
Galkin, V. E., Schmied, W. H., Schraidt, O., Marlovits, T. C. & Egelman, E. H. The structure of the Salmonella typhimurium type III secretion system needle shows divergence from the flagellar system. J. Mol. Biol. 396, 1392–1397 (2010)
Das, R. et al. Simultaneous prediction of protein folding and docking at high resolution. Proc. Natl Acad. Sci. USA 106, 18978–18983 (2009)
Kenjale, R. et al. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J. Biol. Chem. 280, 42929–42937 (2005)
Goldbourt, A., Gross, B. J., Day, L. A. & McDermott, A. E. Filamentous phage studied by magic-angle spinning NMR: resonance assignment and secondary structure of the coat protein in Pf1. J. Am. Chem. Soc. 129, 2338–2344 (2007)
Han, Y. et al. Solid-state NMR studies of HIV-1 capsid protein assemblies. J. Am. Chem. Soc. 132, 1976–1987 (2010)
Jehle, S. et al. Solid-state NMR and SAXS studies provide a structural basis for the activation of αB-crystallin oligomers. Nature Struct. Mol. Biol. 17, 1037–1042 (2010)
Bockmann, A. et al. Characterization of different water pools in solid-state NMR protein samples. J. Biomol. NMR 45, 319–327 (2009)
Fung, B. M., Khitrin, A. K. & Ermolaev, K. An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 142, 97–101 (2000)
Szeverenyi, N. M., Sullivan, M. J. & Maciel, G. E. Observation of spin exchange by two-dimensional Fourier-transform C-13 cross polarization-magic-angle spinning. J. Magn. Reson. 47, 462–475 (1982)
Vranken, W. F. et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696 (2005)
Bradley, P., Misura, K. M. & Baker, D. Toward high-resolution de novo structure prediction for small proteins. Science 309, 1868–1871 (2005)
Acknowledgements
We thank T. C. Marlovits and E. H. Egelman for providing the S. typhimurium T3SS needle cryo-electron microscopy density map; F. DiMaio and J.-P. Demers for discussions; and G. Wolf, B. Angerstein and G. Heim for technical help. This work was supported by the Max Planck Society (to C. Griesinger), the Deutsche Forschungsgemeinschaft (Emmy Noether Fellowship to A. Lange), the Fondation Bettencourt Schueller (to A. Loquet), the National Institutes of Health (1 R01 GM092802-01 to D.B.), EMBO (postdoctoral fellowship to A. Loquet), and the European Union Seventh Framework Program under Grant Agreement 261863 (Bio-NMR).
Author information
Authors and Affiliations
Contributions
A. Loquet performed ssNMR experiments. A. Loquet and A. Lange analysed ssNMR data. N.S. and D.B. performed structure calculations. K.G. and S.B. expressed, purified and polymerized in vitro T3SS needles. R.G. and M.K. performed the in vivo studies. C. Griesinger analysed NMR data. D.R. and C. Goosmann performed electron microscopy studies. A. Loquet and A. Lange wrote the paper; all authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-18, Supplementary Tables 1-3 and Supplementary References. (PDF 7243 kb)
Rights and permissions
About this article
Cite this article
Loquet, A., Sgourakis, N., Gupta, R. et al. Atomic model of the type III secretion system needle. Nature 486, 276–279 (2012). https://doi.org/10.1038/nature11079
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11079
This article is cited by
-
Nonpathogenic Pseudomonas syringae derivatives and its metabolites trigger the plant “cry for help” response to assemble disease suppressing and growth promoting rhizomicrobiome
Nature Communications (2024)
-
Efficient 18.8 T MAS-DNP NMR reveals hidden side chains in amyloid fibrils
Journal of Biomolecular NMR (2023)
-
Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation
Nature Communications (2021)
-
Structural analysis of cross α-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials
Nature Communications (2021)
-
Extensively sparse 13C labeling to simplify solid-state NMR 13C spectra of membrane proteins
Journal of Biomolecular NMR (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.