Abstract
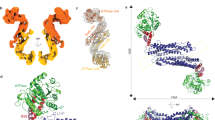
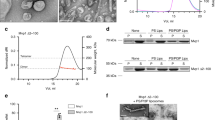
Dynamin-related proteins (DRPs) are multi-domain GTPases that function via oligomerization and GTP-dependent conformational changes to play central roles in regulating membrane structure across phylogenetic kingdoms. How DRPs harness self-assembly and GTP-dependent conformational changes to remodel membranes is not understood. Here we present the crystal structure of an assembly-deficient mammalian endocytic DRP, dynamin 1, lacking the proline-rich domain, in its nucleotide-free state. The dynamin 1 monomer is an extended structure with the GTPase domain and bundle signalling element positioned on top of a long helical stalk with the pleckstrin homology domain flexibly attached on its opposing end. Dynamin 1 dimer and higher order dimer multimers form via interfaces located in the stalk. Analysis of these interfaces provides insight into DRP family member specificity and regulation and provides a framework for understanding the biogenesis of higher order DRP structures and the mechanism of DRP-mediated membrane scission events.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Praefcke, G. J. & McMahon, H. T. The dynamin superfamily: universal membrane tubulation and fission molecules? Nature Rev. Mol. Cell Biol. 5, 133–147 (2004)
Hoppins, S., Lackner, L. & Nunnari, J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76, 751–780 (2007)
Marks, B. et al. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature 410, 231–235 (2001)
Stowell, M. H. B., Marks, B., Wigge, P. & McMahon, H. T. Nucleotide-dependent conformational changes in dynamin: evidence for a mechanochemical molecular spring. Nature Cell Biol. 1, 27–32 (1999)
Mears, J. A. et al. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nature Struct. Mol. Biol. 18, 20–26 (2011)
Warnock, D. E., Hinshaw, J. E. & Schmid, S. L. Dynamin self-assembly stimulates its GTPase activity. J. Biol. Chem. 271, 22310–22314 (1996)
Danino, D., Moon, K. H. & Hinshaw, J. E. Rapid constriction of lipid bilayers by the mechanochemical enzyme dynamin. J. Struct. Biol. 147, 259–267 (2004)
Pucadyil, T. J. & Schmid, S. L. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell 135, 1263–1275 (2008)
Chappie, J. S. et al. An intramolecular signaling element that modulates dynamin function in vitro and in vivo . Mol. Biol. Cell 20, 3561–3571 (2009)
Chappie, J. S., Acharya, S., Leonard, M., Schmid, S. L. & Dyda, F. G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature 465, 435–440 (2010)
Timm, D. et al. Crystal structure of the pleckstrin homology domain from dynamin. Nature Struct. Biol. 1, 782–788 (1994)
Achiriloaie, M., Barylko, B. & Albanesi, J. P. Essential role of the dynamin pleckstrin homology domain in receptor-mediated endocytosis. Mol. Cell. Biol. 19, 1410–1415 (1999)
Ferguson, K. M., Lemmon, M. A., Schlessinger, J. & Sigler, P. B. Crystal structure at 2.2 Å resolution of the pleckstrin homology domain from human dynamin. Cell 79, 199–209 (1994)
Ramachandran, R. et al. Membrane insertion of the pleckstrin homology domain variable loop 1 is critical for dynamin-catalyzed vesicle scission. Mol. Biol. Cell 20, 4630–4639 (2009)
Hinshaw, J. E. & Schmid, S. L. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature 374, 190–192 (1995)
Ingerman, E. et al. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 170, 1021–1027 (2005)
Mears, J. A., Ray, P. & Hinshaw, J. E. A corkscrew model for dynamin constriction. Structure 15, 1190–1202 (2007)
Gao, S. et al. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465, 502–506 (2010)
Low, H. H., Sachse, C., Amos, L. A. & Lowe, J. Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell 139, 1342–1352 (2009)
Low, H. H. & Lowe, J. A bacterial dynamin-like protein. Nature 444, 766–769 (2006)
Prakash, B., Praefcke, G. J., Renault, L., Wittinghofer, A. & Herrmann, C. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature 403, 567–571 (2000)
Ghosh, A., Praefcke, G. J., Renault, L., Wittinghofer, A. & Herrmann, C. How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature 440, 101–104 (2006)
Bian, X. et al. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc. Natl Acad. Sci. USA 108, 3976–3981 (2011)
Byrnes, L. J. & Sondermann, H. Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc. Natl Acad. Sci. USA 108, 2216–2221 (2011)
Zhang, P. & Hinshaw, J. E. Three-dimensional reconstruction of dynamin in the constricted state. Nature Cell Biol. 3, 922–926 (2001)
Siegel, L. M. & Monty, K. J. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta 112, 346–362 (1966)
Ramachandran, R. et al. The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO J. 26, 559–566 (2007)
Gao, S. et al. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465, 502–506 (2010)
Song, B. D., Yarar, D. & Schmid, S. L. An assembly-incompetent mutant establishes a requirement for dynamin self-assembly in clathrin-mediated endocytosis in vivo . Mol. Biol. Cell 15, 2243–2252 (2004)
Reubold, T. F. et al. Crystal structure of the GTPase domain of rat dynamin 1. Proc. Natl Acad. Sci. USA 102, 13093–13098 (2005)
Sweitzer, S. M. & Hinshaw, J. E. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell 93, 1021–1029 (1998)
Bashkirov, P. V. et al. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell 135, 1276–1286 (2008)
Kenniston, J. A. & Lemmon, M. A. Dynamin GTPase regulation is altered by PH domain mutations found in centronuclear myopathy patients. EMBO J. 29, 3054–3067 (2010)
Niemann, H. H., Knetsch, M. L., Scherer, A., Manstein, D. J. & Kull, F. J. Crystal structure of a dynamin GTPase domain in both nucleotide-free and GDP-bound forms. EMBO J. 20, 5813–5821 (2001)
Barylko, B. et al. The proline/arginine-rich domain is a major determinant of dynamin self-activation. Biochemistry 49, 10592–10594 (2010)
Vallis, Y., Wigge, P., Marks, B., Evans, P. R. & McMahon, H. T. Importance of the pleckstrin homology domain of dynamin in clathrin-mediated endocytosis. Curr. Biol. 9, 257–263 (1999)
Durieux, A. C., Prudhon, B., Guicheney, P. & Bitoun, M. Dynamin 2 and human diseases. J. Mol. Med. 88, 339–350 (2010)
Lackner, L. L., Horner, J. S. & Nunnari, J. Mechanistic analysis of a dynamin effector. Science 325, 874–877 (2009)
Gandre-Babbe, S. & van der Bliek, A. M. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 19, 2402–2412 (2008)
Kosaka, T. & Ikeda, K. Possible temperature-dependent blockage of synaptic vesicle recycling induced by a single gene mutation in Drosophila . J. Neurobiol. 14, 207–225 (1983)
Ramaswami, M., Rao, S., van der Bliek, A., Kelly, R. B. & Krishnan, K. S. Genetic studies on dynamin function in Drosophila . J. Neurogenet. 9, 73–87 (1993)
Narayanan, R., Leonard, M., Song, B. D., Schmid, S. L. & Ramaswami, M. An internal GAP domain negatively regulates presynaptic dynamin in vivo: a two-step model for dynamin function. J. Cell Biol. 169, 117–126 (2005)
Ingerman, E. & Nunnari, J. A continuous, regenerative coupled GTPase assay for dynamin-related proteins. Methods Enzymol. 404, 611–619 (2005)
Acknowledgements
The authors would like to express thanks to I. Stokes-Rees for assistance with the Wide Space Molecular Replacement, J. Holton for advice and assistance with data collection and H. McMahon for the kind gift of the dynamin clone. We would also like to thank J. Al-Bassam, J. Chappie, A. McCoy, S. Harrison, D. Owen, A. Fisher, E. Baldwin, L. Lackner, G. Adamson, N. Varlakhanova and members of the Nunnari lab for extensive discussions. S.J. is a Damon Runyon Cancer Research Foundation Fellow supported by the Howard Hughes Medical Institute (DRG-2004-09). This work was supported by an American Heart Innovator Award and NIH grants (R01GM062942S1 and R01GM097432) to J.N. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under Contract No. DE-AC02-05CH11231.
Author information
Authors and Affiliations
Contributions
M.G.J.F. purified, biochemically characterized and crystallized the Dyn1 G397D ΔPRD. M.G.J.F. collected X-ray diffraction data and M.G.J.F. and S.J. solved the structure. M.G.J.F. and J.N. designed experiments and M.G.J.F., S.J. and J.N. interpreted data and prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-5 with legends, Supplementary Methods, Supplementary Tables 1-3 and Supplementary References. This file was corrected on 07 October 2011 to change the Protein names from rDyn1 to Dyn1 and Dynamin-1 to Dynamin respectively. (PDF 2774 kb)
Rights and permissions
About this article
Cite this article
Ford, M., Jenni, S. & Nunnari, J. The crystal structure of dynamin. Nature 477, 561–566 (2011). https://doi.org/10.1038/nature10441
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10441
This article is cited by
-
A deep intronic variant in DNM1 in a patient with developmental and epileptic encephalopathy creates a splice acceptor site and affects only transcript variants including exon 10a
neurogenetics (2023)
-
Function and regulation of the divisome for mitochondrial fission
Nature (2021)
-
Drp1 is widely, yet heterogeneously, distributed in the mouse central nervous system
Molecular Brain (2020)
-
The cell biology of mitochondrial membrane dynamics
Nature Reviews Molecular Cell Biology (2020)
-
Dynamin-2 R465W mutation induces long range perturbation in highly ordered oligomeric structures
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.