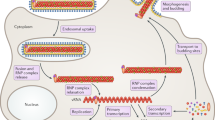
Abstract
Ebola virus (EboV) is a highly pathogenic enveloped virus that causes outbreaks of zoonotic infection in Africa. The clinical symptoms are manifestations of the massive production of pro-inflammatory cytokines in response to infection1 and in many outbreaks, mortality exceeds 75%. The unpredictable onset, ease of transmission, rapid progression of disease, high mortality and lack of effective vaccine or therapy have created a high level of public concern about EboV2. Here we report the identification of a novel benzylpiperazine adamantane diamide-derived compound that inhibits EboV infection. Using mutant cell lines and informative derivatives of the lead compound, we show that the target of the inhibitor is the endosomal membrane protein Niemann–Pick C1 (NPC1). We find that NPC1 is essential for infection, that it binds to the virus glycoprotein (GP), and that antiviral compounds interfere with GP binding to NPC1. Combined with the results of previous studies of GP structure and function, our findings support a model of EboV infection in which cleavage of the GP1 subunit by endosomal cathepsin proteases removes heavily glycosylated domains to expose the amino-terminal domain3,4,5,6,7, which is a ligand for NPC1 and regulates membrane fusion by the GP2 subunit8. Thus, NPC1 is essential for EboV entry and a target for antiviral therapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Zampieri, C. A., Sullivan, N. J. & Nabel, G. J. Immunopathology of highly virulent pathogens: insights from Ebola virus. Nature Immunol. 8, 1159–1164 (2007)
Geisbert, T. W. & Jahrling, P. B. Exotic emerging viral diseases: progress and challenges. Nature Med. 10, S110–S121 (2004)
Chandran, K., Sullivan, N. J., Felbor, U., Whelan, S. P. & Cunningham, J. M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308, 1643–1645 (2005)
Schornberg, K. et al. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 80, 4174–4178 (2006)
Lee, J. E. et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454, 177–182 (2008)
Dube, D. et al. The primed ebolavirus glycoprotein (19-kilodalton GP1,2): Sequence and residues critical for host cell binding. J. Virol. 83, 2883–2891 (2009)
Hood, C. L. et al. Biochemical and structural characterization of cathepsin L-processed Ebola virus glycoprotein: Implications for viral entry and immunogenicity. J. Virol. 84, 2972–2982 (2010)
Harrison, S. C. Viral membrane fusion. Nature Struct. Mol. Biol. 15, 690–698 (2008)
Wong, A., Sandesara, R., Mulherkar, N., Whelan, S. & Chandran, K. A forward genetic strategy reveals destabilizing mutations in the ebolavirus glycoprotein that alter its protease dependence during cell entry. J. Virol. 84, 163–175 (2010)
Kolter, T. & Sandhoff, K. Lysosomal degradation of membrane lipids. FEBS Lett. 584, 1700–1712 (2010)
Du, X. et al. A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J. Cell Biol. 192, 121–135 (2011)
Chevallier, J. et al. Lysobisphosphatidic acid controls endosomal cholesterol levels. J. Biol. Chem. 283, 27871–27880 (2008)
Ko, D. C., Gordon, M. D., Jin, J. Y. & Scott, M. P. Dynamic movements of organelles containing Niemann-Pick C1 protein: NPC1 involvement in late endocytic events. Mol. Biol. Cell 12, 601–614 (2001)
Millard, E. E. et al. The sterol-sensing domain of the Niemann-Pick C1 (NPC1) protein regulates trafficking of low density lipoprotein cholesterol. J. Biol. Chem. 280, 28581–28590 (2005)
Ohgami, N. et al. Binding between the Niemann–Pick C1 protein and a photoactivatable cholesterol analog requires a functional sterol-sensing domain. Proc. Natl Acad. Sci. USA 101, 12473–12478 (2004)
Towner, J. S. et al. Newly discovered Ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 4, e1000212 (2008)
Kuhn, J. H. et al. Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J. Biol. Chem. 281, 15951–15958 (2006)
Kaletsky, R. L., Simmons, G. & Bates, P. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J. Virol. 81, 13378–13384 (2007)
Brindley, M. A. et al. Ebola virus glycoprotein 1: Identification of residues important for binding and postbinding events. J. Virol. 81, 7702–7709 (2007)
Dube, D. et al. Cell adhesion-dependent membrane trafficking of a binding partner for the ebolavirus glycoprotein is a determinant of viral entry. Proc. Natl Acad. Sci. USA 107, 16637–16642 (2010)
Ban, H. S. et al. Identification of HSP60 as a primary target of o-carboranylphenylphenoxyacetanilide, an HIF-1α inhibitor. J. Am. Chem. Soc. 132, 11870–11871 (2010)
Sobo, K. et al. Late endosomal cholesterol accumulation leads to impaired intra-endosomal trafficking. PLoS ONE 2, e851 (2007)
Huynh, K. K., Gershenzon, E. & Grinstein, S. Cholesterol accumulation by macrophages impairs phagosome maturation. J. Biol. Chem. 283, 35745–35755 (2008)
Saeed, M. F., Kolokoltsov, A. A., Albrecht, T. & Davey, R. A. Cellular entry of Ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 6, e1001110 (2010)
Nanbo, A. et al. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 6, e1001121 (2010)
Gelsthorpe, M. E. et al. Niemann-Pick type C1 I1061T mutant encodes a functional protein that is selected for endoplasmic reticulum-associated degradation due to protein misfolding. J. Biol. Chem. 283, 8229–8236 (2008)
Millard, E. E., Srivastava, K., Traub, L. M., Schaffer, J. E. & Ory, D. S. Niemann-pick type C1 (NPC1) overexpression alters cellular cholesterol homeostasis. J. Biol. Chem. 275, 38445–38451 (2000)
Soneoka, Y. et al. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23, 628–633 (1995)
Towner, J. S. et al. Generation of eGFP expressing recombinant Zaire ebolavirus for analysis of early pathogenesis events and high-throughput antiviral drug screening. Virology 332, 20–27 (2005)
Weidmann, M., Mühlberger, E. & Hufert, F. T. Rapid detection protocol for filoviruses. J. Clin. Virol. 30, 94–99 (2004)
Acknowledgements
We thank B. Considine, A. Nilsson and S. Wilkes for assistance, S. Chiang for critical reading of the manuscript, G. Beltz, N. Gray, S. Grinstein, Y. Iannou, R. Infante, J. Kornhuber, F. Sharom and S. Whelan for discussion. This work was supported by grants from U54 AI057159, R01 CA104266 to J.C., PIDS-Sanofi-Pasteur Fellowship, K12-HD052896 and 5K08AI079381 to J.M., 5-T32- HL007623 to A.B., and fellowship from Fonds de la Recherche en Santé du Québec to M.C.; C.M.F. was supported by the Postgraduate Research Participation Program at the US Army Medical Research and Material Command administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and USAMRMC.
Author information
Authors and Affiliations
Contributions
M.C., J.M., T.R. and A.B. equally contributed to this work. K.C. and T.R. performed the inhibitor screen. K.L. synthesized and purified 3.0 analogues and T.R. tested them. T.R., A.B., J.M., Q.L. and M.C. carried out infection assays with pseudotyped viruses. A.B. performed microscopy. J.M. purified recombinant glycoprotein. M.C. and J.M. designed and performed binding assays. M.C. performed immunoprecipitation. D.O. provided NPC1 constructs, antibodies and CHO cell lines. Ebola virus infections were performed in the lab of L.H. by C.M.F.; J.C. supervised the project and wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-8 with legends and Supplementary Text and Data detailing the synthesis and characterization of the novel chemicals used in the manuscript. (PDF 4560 kb)
Rights and permissions
About this article
Cite this article
Côté, M., Misasi, J., Ren, T. et al. Small molecule inhibitors reveal Niemann–Pick C1 is essential for Ebola virus infection. Nature 477, 344–348 (2011). https://doi.org/10.1038/nature10380
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10380
This article is cited by
-
Tubeimosides are pan-coronavirus and filovirus inhibitors that can block their fusion protein binding to Niemann-Pick C1
Nature Communications (2024)
-
An overview of the role of Niemann-pick C1 (NPC1) in viral infections and inhibition of viral infections through NPC1 inhibitor
Cell Communication and Signaling (2023)
-
The roles of different microRNAs in the regulation of cholesterol in viral hepatitis
Cell Communication and Signaling (2023)
-
Identification of CCZ1 as an essential lysosomal trafficking regulator in Marburg and Ebola virus infections
Nature Communications (2023)
-
Longitudinal in vivo imaging of acute neuropathology in a monkey model of Ebola virus infection
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.