Abstract
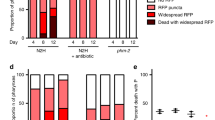
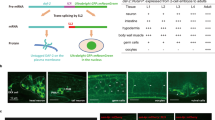
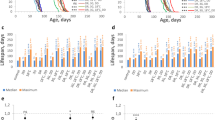
Overexpression of sirtuins (NAD+-dependent protein deacetylases) has been reported to increase lifespan in budding yeast (Saccharomyces cerevisiae), Caenorhabditis elegans and Drosophila melanogaster1,2,3. Studies of the effects of genes on ageing are vulnerable to confounding effects of genetic background4. Here we re-examined the reported effects of sirtuin overexpression on ageing and found that standardization of genetic background and the use of appropriate controls abolished the apparent effects in both C. elegans and Drosophila. In C. elegans, outcrossing of a line with high-level sir-2.1 overexpression1 abrogated the longevity increase, but did not abrogate sir-2.1 overexpression. Instead, longevity co-segregated with a second-site mutation affecting sensory neurons. Outcrossing of a line with low-copy-number sir-2.1 overexpression2 also abrogated longevity. A Drosophila strain with ubiquitous overexpression of dSir2 using the UAS-GAL4 system was long-lived relative to wild-type controls, as previously reported3, but was not long-lived relative to the appropriate transgenic controls, and nor was a new line with stronger overexpression of dSir2. These findings underscore the importance of controlling for genetic background and for the mutagenic effects of transgene insertions in studies of genetic effects on lifespan. The life-extending effect of dietary restriction on ageing in Drosophila has also been reported to be dSir2 dependent3. We found that dietary restriction increased fly lifespan independently of dSir2. Our findings do not rule out a role for sirtuins in determination of metazoan lifespan, but they do cast doubt on the robustness of the previously reported effects of sirtuins on lifespan in C. elegans and Drosophila.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Tissenbaum, H. A. & Guarente, L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410, 227–230 (2001)
Viswanathan, M., Kim, S. K., Berdichevsky, A. & Guarente, L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev. Cell 9, 605–615 (2005)
Rogina, B. & Helfand, S. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc. Natl Acad. Sci. USA 101, 15998–16003 (2004)
Partridge, L. & Gems, D. Benchmarks for ageing studies. Nature 450, 165–167 (2007)
Kaeberlein, M., McVey, M. & Guarente, L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13, 2570–2580 (1999)
Berdichevsky, A., Viswanathan, M., Horvitz, H. R. & Guarente, L. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell 125, 1165–1177 (2006)
Guarente, L. Sirtuins in aging and disease. Cold Spring Harb. Symp. Quant. Biol. 72, 483–488 (2007)
Mair, W. & Dillin, A. Aging and survival: the genetics of life span extension by dietary restriction. Annu. Rev. Biochem. 77, 727–754 (2008)
Lin, S., Defossez, P. & Guarente, L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289, 2126–2128 (2000)
Wang, Y. & Tissenbaum, H. A. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech. Ageing Dev. 127, 48–56 (2006)
Haigis, M. C. & Sinclair, D. A. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5, 253–295 (2010)
Garber, K. A mid-life crisis for aging theory. Nature Biotechnol. 26, 371–374 (2008)
Kaeberlein, M. Lessons on longevity from budding yeast. Nature 464, 513–519 (2010)
Kenyon, C. The genetics of ageing. Nature 464, 504–512 (2010)
Howitz, K. T. et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425, 191–196 (2003)
Milne, J. et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450, 712–716 (2007)
Kaeberlein, M. et al. Substrate-specific activation of sirtuins by resveratrol. J. Biol. Chem. 280, 17038–17045 (2005)
Borra, M., Smith, B. & Denu, J. Mechanism of human SIRT1 activation by resveratrol. J. Biol. Chem. 280, 17187–17195 (2005)
Beher, D. et al. Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem. Biol. Drug Des. 74, 619–624 (2009)
Pacholec, M. et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J. Biol. Chem. 285, 8340–8351 (2010)
Hedgecock, E., Culotti, J., Thomson, J. & Perkins, L. Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev. Biol. 111, 158–170 (1985)
Apfeld, J. & Kenyon, C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature 402, 804–809 (1999)
Brand, A. & Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993)
Parker, J. A. et al. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nature Genet. 37, 349–350 (2005)
Patel, D. S. et al. Clustering of genetically defined allele classes in the Caenorhabditis elegans DAF-2 insulin/IGF-1 receptor. Genetics 178, 931–946 (2008)
Wood, J. et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 430, 686–689 (2004)
Bass, T., Weinkove, D., Houthoofd, K., Gems, D. & Partridge, L. Effects of resveratrol on lifespan in Drosophila melanogaster and Caenorhabditis elegans. Mech. Ageing Dev. 128, 546–552 (2007)
Gems, D. et al. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics 150, 129–155 (1998)
Borra, M. T. & Denu, J. M. Quantitative assays for characterization of the Sir2 family of NAD+-dependent deacetylases. Methods Enzymol. 376, 171–187 (2003)
Grandison, R. C., Wong, R., Bass, T. M., Partridge, L. & Piper, M. D. Effect of a standardised dietary restriction protocol on multiple laboratory strains of Drosophila melanogaster. PLoS ONE 4, e4067 (2009)
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974)
Sulston, J. & Hodgkin, J. in The Nematode Caenorhabditis elegans (ed. Wood, W. B. ) 587–606 (Cold Spring Harbor, 1988)
Kamath, R. et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 (2003)
Greiss, S., Hall, J., Ahmed, S. & Gartner, A. C. elegans SIR-2.1 translocation is linked to a proapoptotic pathway parallel to cep-1/p53 during DNA damage-induced apoptosis. Genes Dev. 22, 2831–2842 (2008)
Newman, B. L., Lundblad, J. R., Chen, Y. & Smolik, S. M. A Drosophila homologue of Sir2 modifies position-effect variegation but does not affect life span. Genetics 162, 1675–1685 (2002)
Astrom, S. U., Cline, T. W. & Rine, J. The Drosophila melanogaster sir2+gene is nonessential and has only minor effects on position-effect variegation. Genetics 163, 931–937 (2003)
Acknowledgements
We thank A. Gartner for providing an antibody against C. elegans SIR-2.1; D. Chen, P. Kapahi, S. Pletcher and D. Skorupa for providing data; S. S. Lee for permission to cite unpublished results; S. Helfand and J. Rine for providing fly strains; W. Mair for performing preliminary trials and R. Baumeister for useful discussion. Some nematode strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. We acknowledge funding from the European Union (FP6-036894 to C.B., D.G., L.P. and S.V. and FP6-518230 to D.G. and C.S.), the Hungarian Science Foundation and Norway Grants (NNF-78794 to C.S.), INSERM and ANR, Paris (R.V., A.-M.O. and C.N.), the National Institutes of Health (CA129132 to A.B., R01AG031108 to M.K. and T32AG000057 to G.L.S.) and the Wellcome Trust (Strategic Award to C.A., F.C., D.G., L.P. and M.R.). C.S. is a Bolyai Research Scholar of the Hungarian Academy of Sciences and M.K. is an Ellison Medical Foundation New Scholar in Aging.
Author information
Authors and Affiliations
Contributions
The project was conceived by D.G. and L.P. and the experiments were designed by A.B., C.B., F.C., D.G., K.H., M.K., J.J.M., C.N., L.P., C.S. and S.V. The experiments were performed and analysed by C.A., D.A., C.B., F.C., J.J.M., M.G., M.H., A.-M.O., M.D.P., M.R., G.L.S., M.S., G.V., R.P.V.-M., S.V. and V.L. The manuscript was written by C.B., F.C., D.G., L.P. and S.V.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-12 with legends, Supplementary Tables 1-7 and Supplementary References. (PDF 976 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Burnett, C., Valentini, S., Cabreiro, F. et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature 477, 482–485 (2011). https://doi.org/10.1038/nature10296
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10296
This article is cited by
-
Molecular hallmarks of ageing in amyotrophic lateral sclerosis
Cellular and Molecular Life Sciences (2024)
-
Sirtuins and Aging
Neuroscience and Behavioral Physiology (2023)
-
Resveratrol and Sir2 Reverse Sleep and Memory Defects Induced by Amyloid Precursor Protein
Neuroscience Bulletin (2023)
-
Fatty acid tryptamide from cacao elongates Drosophila melanogaster lifespan with sirtuin-dependent heat shock protein expression
Scientific Reports (2022)
-
Molecular mechanisms of dietary restriction promoting health and longevity
Nature Reviews Molecular Cell Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.