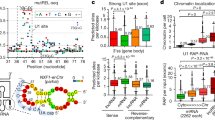
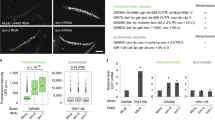
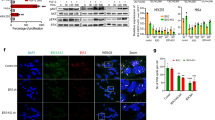
Abstract
Non-coding (nc)RNAs are key players in numerous biological processes such as gene regulation, chromatin domain formation and genome stability1,2. Large ncRNAs interact with histone modifiers3,4,5 and are involved in cancer development6, X-chromosome inactivation7 and autosomal gene imprinting8. However, despite recent evidence showing that pervasive transcription is more widespread than previously thought9, only a few examples mediating gene regulation in eukaryotes have been described10. In Saccharomyces cerevisiae, the bona-fide regulatory ncRNAs are destabilized by the Xrn1 5′–3′ RNA exonuclease11,12 (also known as Kem1), but the genome-wide characterization of the entire regulatory ncRNA family remains elusive. Here, using strand-specific RNA sequencing (RNA-seq), we identify a novel class of 1,658 Xrn1-sensitive unstable transcripts (XUTs) in which 66% are antisense to open reading frames. These transcripts are polyadenylated and RNA polymerase II (RNAPII)-dependent. The majority of XUTs strongly accumulate in lithium-containing media, indicating that they might have a role in adaptive responses to changes in growth conditions. Notably, RNAPII chromatin immunoprecipitation followed by DNA sequencing (ChIP-seq) analysis of Xrn1-deficient strains revealed a significant decrease of RNAPII occupancy over 273 genes with antisense XUTs. These genes show an unusual bias for H3K4me3 marks and require the Set1 histone H3 lysine 4 methyl-transferase for silencing. Furthermore, abolishing H3K4me3 triggers the silencing of other genes with antisense XUTs, supporting a model in which H3K4me3 antagonizes antisense ncRNA repressive activity. Our results demonstrate that antisense ncRNA-mediated regulation is a general regulatory pathway for gene expression in S. cerevisiae.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Sequence Read Archive
Data deposits
Sequence data are publicly available at NCBI Sequence Read Archive under accession number SRA030505 and at http://vm-gb.curie.fr/XUT/index.htm.
References
Bernstein, E. & Allis, C. D. RNA meets chromatin. Genes Dev. 19, 1635–1655 (2005)
Moazed, D. Small RNAs in transcriptional gene silencing and genome defence. Nature 457, 413–420 (2009)
Swiezewski, S., Liu, F., Magusin, A. & Dean, C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462, 799–802 (2009)
Yu, W. et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451, 202–206 (2008)
Yap, K. L. et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a . Mol. Cell 38, 662–674 (2010)
Huarte, M. & Rinn, J. L. Large non-coding RNAs: missing links in cancer? Hum. Mol. Genet. 19, R152–R161 (2010)
Chow, J. & Heard, E. X inactivation and the complexities of silencing a sex chromosome. Curr. Opin. Cell Biol. 21, 359–366 (2009)
Nagano, T. et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717–1720 (2008)
Amaral, P. P., Dinger, M. E., Mercer, T. R. & Mattick, J. S. The eukaryotic genome as an RNA machine. Science 319, 1787–1789 (2008)
Berretta, J. & Morillon, A. Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Rep. 10, 973–982 (2009)
Camblong, J. et al. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae . Genes Dev. 23, 1534–1545 (2009)
Berretta, J., Pinskaya, M. & Morillon, A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae . Genes Dev. 22, 615–626 (2008)
Matsuda, E. & Garfinkel, D. J. Posttranslational interference of Ty1 retrotransposition by antisense RNAs. Proc. Natl Acad. Sci. USA 106, 15657–15662 (2009)
Aravind, L., Watanabe, H., Lipman, D. J. & Koonin, E. V. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc. Natl Acad. Sci. USA 97, 11319–11324 (2000)
Jacquier, A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nature Rev. Genet. 10, 833–844 (2009)
Long, R. M. & McNally, M. T. mRNA decay: X (XRN1) marks the spot. Mol. Cell 11, 1126–1128 (2003)
Nagalakshmi, U. et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320, 1344–1349 (2008)
Neil, H. et al. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457, 1038–1042 (2009)
Yassour, M. et al. Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol. 11, R87 (2010)
Chernyakov, I., Whipple, J. M., Kotelawala, L., Grayhack, E. J. & Phizicky, E. M. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′–3′ exonucleases Rat1 and Xrn1. Genes Dev. 22, 1369–1380 (2008)
Fatica, A., Morlando, M. & Bozzoni, I. Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J. 19, 6218–6229 (2000)
Xu, Z. et al. Bidirectional promoters generate pervasive transcription in yeast. Nature 457, 1033–1037 (2009)
Thompson, D. M. & Parker, R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae . Mol. Cell. Biol. 27, 92–101 (2007)
Wyers, F. et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121, 725–737 (2005)
Dichtl, B., Stevens, A. & Tollervey, D. Lithium toxicity in yeast is due to the inhibition of RNA processing enzymes. EMBO J. 16, 7184–7195 (1997)
Johnson, A. W. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 17, 6122–6130 (1997)
Pinskaya, M. & Morillon, A. Histone H3 lysine 4 di-methylation: a novel mark for transcriptional fidelity? Epigenetics 4, 302–306 (2009)
Pokholok, D. K. et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517–527 (2005)
Kirmizis, A. et al. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449, 928–932 (2007)
Longtine, M. S. et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast 14, 953–961 (1998)
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression. Bioinformatics 26, 139–140 (2010)
Dichtl, B., Aasland, R. & Keller, W. Functions for S. cerevisiae Swd2p in 3′ end formation of specific mRNAs and snoRNAs and global histone 3 lysine 4 methylation. RNA 10, 965–977 (2004)
Pinskaya, M., Gourvennec, S. & Morillon, A. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 28, 1697–1707 (2009)
Camblong, J., Iglesias, N., Fickentscher, C., Dieppois, G. & Stutz, F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae . Cell 131, 706–717 (2007)
Nonet, M., Scafe, C., Sexton, J. & Young, R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol. 7, 1602–1611 (1987)
Acknowledgements
We thank B. Séraphin, L. Bénard and J. O’Sullivan for support and advice; B. Dichtl for insights into lithium treatment data normalization; M. Wéry and A. Taddei for helpful discussions; A. Johnson, T. Kouzarides and V. Géli for generous gift of plasmids and strains. Special thanks to M. Descrimes, C. Jubin and S. Lair for technical assistance. We thank L. Steinmetz and M. Chodder for sharing unpublished results. E.L.V.D. benefits from an FRM fellowship. This work has benefited from facilities and expertise of the IMAGIF sequencing platform (Centre de Recherche de Gif). This work was financially supported by the Canceropole Ile de France, the ANR “REGULncRNA” and ERC “EPIncRNA” starting grant.
Author information
Authors and Affiliations
Contributions
E.L.V.D. performed molecular biology experiments, RNA-seq, ChIP-seq libraries and sequencing on the ILLUMINA platform. C.L.C., Y.D.-C., M.S. and C.T. performed statistical and bioinformatic analyses. S.G., M.K., V.R. and C.B. provided technical assistance to molecular biology experiments. A.M., C.T., E.L.V.D. and C.L.C. designed the experiments. P.L.-N. and S.L. performed RNA-seq libraries and NGS sequencing on the SOLiD platform; A.N. managed sequencing on the SOLiD platform. E.L.V.D., C.L.C. and A.N. contributed to the writing. C.T. and A.M. wrote the paper. C.T. and A.M. planned the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Figures 1-20 with legends, Supplementary References and Supplementary Tables 1-3. (PDF 1807 kb)
Rights and permissions
About this article
Cite this article
van Dijk, E., Chen, C., d’Aubenton-Carafa, Y. et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475, 114–117 (2011). https://doi.org/10.1038/nature10118
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10118
This article is cited by
-
CTCF blocks antisense transcription initiation at divergent promoters
Nature Structural & Molecular Biology (2022)
-
Uncovering de novo gene birth in yeast using deep transcriptomics
Nature Communications (2021)
-
Global view on the metabolism of RNA poly(A) tails in yeast Saccharomyces cerevisiae
Nature Communications (2021)
-
Global identification of a marine diatom long noncoding natural antisense transcripts (NATs) and their response to phosphate fluctuations
Scientific Reports (2020)
-
UNAGI: an automated pipeline for nanopore full-length cDNA sequencing uncovers novel transcripts and isoforms in yeast
Functional & Integrative Genomics (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.