Abstract
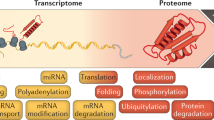
Gene expression is a multistep process that involves the transcription, translation and turnover of messenger RNAs and proteins. Although it is one of the most fundamental processes of life, the entire cascade has never been quantified on a genome-wide scale. Here we simultaneously measured absolute mRNA and protein abundance and turnover by parallel metabolic pulse labelling for more than 5,000 genes in mammalian cells. Whereas mRNA and protein levels correlated better than previously thought, corresponding half-lives showed no correlation. Using a quantitative model we have obtained the first genome-scale prediction of synthesis rates of mRNAs and proteins. We find that the cellular abundance of proteins is predominantly controlled at the level of translation. Genes with similar combinations of mRNA and protein stability shared functional properties, indicating that half-lives evolved under energetic and dynamic constraints. Quantitative information about all stages of gene expression provides a rich resource and helps to provide a greater understanding of the underlying design principles.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
13 February 2013
Nature 473, 337–342 (2011); doi:10.1038/nature10098 Mark Biggin of the Lawrence Berkeley National Laboratory contacted us, noting that our mass-spectrometry-based protein copy number estimates are lower than several literature-based values. We therefore re-analysed the scripts used for data processing, and found a scaling error that occurred during the conversion of normalized protein intensity values into absolute copy number estimates.
References
Ben-Tabou de-Leon, S. & Davidson, E. H. Modeling the dynamics of transcriptional gene regulatory networks for animal development. Dev. Biol. 325, 317–328 (2009)
Komili, S. & Silver, P. A. Coupling and coordination in gene expression processes: a systems biology view. Nature Rev. Genet. 9, 38–48 (2008)
de Sousa Abreu, R., Penalva, L. O., Marcotte, E. M. & Vogel, C. Global signatures of protein and mRNA expression levels. Mol. Biosyst. 5, 1512–1526 (2009)
Maier, T., Guell, M. & Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 583, 3966–3973 (2009)
Belle, A., Tanay, A., Bitincka, L., Shamir, R. & O’Shea, E. K. Quantification of protein half-lives in the budding yeast proteome. Proc. Natl Acad. Sci. USA 103, 13004–13009 (2006)
Yang, E. et al. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res. 13, 1863–1872 (2003)
Yen, H. C., Xu, Q., Chou, D. M., Zhao, Z. & Elledge, S. J. Global protein stability profiling in mammalian cells. Science 322, 918–923 (2008)
Gouw, J. W., Krijgsveld, J. & Heck, A. J. Quantitative proteomics by metabolic labeling of model organisms. Mol. Cell. Proteomics 9, 11–24 (2010)
Beynon, R. J. & Pratt, J. M. Metabolic labeling of proteins for proteomics. Mol. Cell. Proteomics 4, 857–872 (2005)
Friedel, C. C., Dolken, L., Ruzsics, Z., Koszinowski, U. H. & Zimmer, R. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res. 37, e115 (2009)
Mann, M. Functional and quantitative proteomics using SILAC. Nature Rev. Mol. Cell Biol. 7, 952–958 (2006)
Doherty, M. K., Hammond, D. E., Clague, M. J., Gaskell, S. J. & Beynon, R. J. Turnover of the human proteome: determination of protein intracellular stability by dynamic SILAC. J. Proteome Res. 8, 104–112 (2009)
Milner, E., Barnea, E., Beer, I. & Admon, A. The turnover kinetics of major histocompatibility complex peptides of human cancer cells. Mol. Cell. Proteomics 5, 357–365 (2006)
Lam, Y. W., Lamond, A. I., Mann, M. & Andersen, J. S. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr. Biol. 17, 749–760 (2007)
Schwanhäusser, B., Gossen, M., Dittmar, G. & Selbach, M. Global analysis of cellular protein translation by pulsed SILAC. Proteomics 9, 205–209 (2009)
Selbach, M. et al. Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 (2008)
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnol. 26, 1367–1372 (2008)
Price, J. C., Guan, S., Burlingame, A., Prusiner, S. B. & Ghaemmaghami, S. Analysis of proteome dynamics in the mouse brain. Proc. Natl Acad. Sci. USA 107, 14508–14513 (2010)
Wu, C. C., MacCoss, M. J., Howell, K. E., Matthews, D. E. & Yates, J. R., III Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal. Chem. 76, 4951–4959 (2004)
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods 5, 621–628 (2008)
Lu, P., Vogel, C., Wang, R., Yao, X. & Marcotte, E. M. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nature Biotechnol. 25, 117–124 (2007)
Malmström, J. et al. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans . Nature 460, 762–765 (2009)
Vogel, C. et al. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol. Syst. Biol. 6, 400 (2010)
Geiss, G. K. et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nature Biotechnol. 26, 317–325 (2008)
Darzacq, X. et al. In vivo dynamics of RNA polymerase II transcription. Nature Struct. Mol. Biol. 14, 796–806 (2007)
Arava, Y., Boas, F. E., Brown, P. O. & Herschlag, D. Dissecting eukaryotic translation and its control by ribosome density mapping. Nucleic Acids Res. 33, 2421–2432 (2005)
Wu, G., Nie, L. & Zhang, W. Integrative analyses of posttranscriptional regulation in the yeast Saccharomyces cerevisiae using transcriptomic and proteomic data. Curr. Microbiol. 57, 18–22 (2008)
Kirkpatrick, D. S., Denison, C. & Gygi, S. P. Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nature Cell Biol. 7, 750–757 (2005)
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998)
King, R. W., Deshaies, R. J., Peters, J. M. & Kirschner, M. W. How proteolysis drives the cell cycle. Science 274, 1652–1659 (1996)
Hao, S. & Baltimore, D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nature Immunol. 10, 281–288 (2009)
Legewie, S., Herzel, H., Westerhoff, H. V. & Bluthgen, N. Recurrent design patterns in the feedback regulation of the mammalian signalling network. Mol. Syst. Biol. 4, 190 (2008)
Wagner, A. Energy constraints on the evolution of gene expression. Mol. Biol. Evol. 22, 1365–1374 (2005)
Pedraza, J. M. & Paulsson, J. Effects of molecular memory and bursting on fluctuations in gene expression. Science 319, 339–343 (2008)
Mittal, N., Roy, N., Babu, M. M. & Janga, S. C. Dissecting the expression dynamics of RNA-binding proteins in posttranscriptional regulatory networks. Proc. Natl Acad. Sci. USA 106, 20300–20305 (2009)
Hogan, D. J., Riordan, D. P., Gerber, A. P., Herschlag, D. & Brown, P. O. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 6, e255 (2008)
Hentze, M. W., Muckenthaler, M. U. & Andrews, N. C. Balancing acts: molecular control of mammalian iron metabolism. Cell 117, 285–297 (2004)
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009)
Churchman, L. S. & Weissman, J. S. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469, 368–373 (2011)
Gebauer, F. & Hentze, M. W. Molecular mechanisms of translational control. Nature Rev. Mol. Cell Biol. 5, 827–835 (2004)
Sonenberg, N. & Hinnebusch, A. G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 (2009)
Lu, R. et al. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature 462, 358–362 (2009)
Rosenfeld, N., Elowitz, M. B. & Alon, U. Negative autoregulation speeds the response times of transcription networks. J. Mol. Biol. 323, 785–793 (2002)
Rabani, M. et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nature Biotechnol. 10.1038/nbt.1861 (24 April 2011).
Acknowledgements
We thank N. Rajewsky and L. Dölken for fruitful discussions and C. Sommer for technical assistance. M.S. and W.C. are supported by the Helmholtz Association, the German Ministry of Education and Research (BMBF) and the Senate of Berlin by funds aimed at establishing the Berlin Institute of Medical Systems Biology (BIMSB) (grant number 315362A). J.W. is supported by the ForSys-programme of the German Ministry of Education and Research (grant number 315289); D.B. by the Helmholtz Alliance on Systems Biology/MSBN; and N.L. by the China Scholarship Council CSC.
Author information
Authors and Affiliations
Contributions
M.S. conceived, designed and supervised the experiments. B.S. performed wet-lab experiments, mass spectrometry and proteomic data analysis. D.B. and J.W. developed and employed the mathematical model. N.L. performed RNA-seq experiments. W.C. designed and supervised RNA-seq experiments. B.S., D.B., J.S., W.C. and M.S. analysed genome-wide data. G.D. helped in cycloheximide chase experiments and data analysis. B.S., D.B., J.S., J.W., W.C. and M.S. interpreted the data. M.S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-12 with legends. This file was replaced on 13 February 2013 - see Selbach 11848 corrigendum for details. (PDF 2430 kb)
Supplementary Methods
This file contains Supplementary Methods and Data, Supplementary Figures 1-4 with legends and additional references. (PDF 739 kb)
Supplementary Table 1
This table displays an overview of data reproducibility. This file was replaced on 13 February 2013 - see Selbach 11848 corrigendum for details. (XLS 31 kb)
Supplementary Table 2
This table displays categories enriched in bins of genes with the specified combinations of protein and mRNA half-lives. (XLS 72 kb)
Supplementary Table 3
This table displays protein and mRNA copy numbers, half-lives, transcription rates and translation rate constants in mouse fibroblasts (NIH 3T3). This file was replaced on 13 February 2013 - see Selbach 11848 corrigendum for details. (XLS 3314 kb)
Rights and permissions
About this article
Cite this article
Schwanhäusser, B., Busse, D., Li, N. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). https://doi.org/10.1038/nature10098
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature10098
This article is cited by
-
Decoding of the surfaceome and endocytome in primary glioblastoma cells identifies potential target antigens in the hypoxic tumor niche
Acta Neuropathologica Communications (2024)
-
Proteomic analysis of breast cancer based on immune subtypes
Clinical Proteomics (2024)
-
Phase separation as a possible mechanism for dosage sensitivity
Genome Biology (2024)
-
METTL3 drives NSCLC metastasis by enhancing CYP19A1 translation and oestrogen synthesis
Cell & Bioscience (2024)
-
Ribosome profiling: a powerful tool in oncological research
Biomarker Research (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.