Abstract
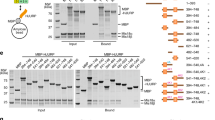
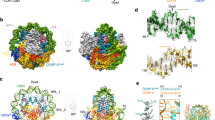
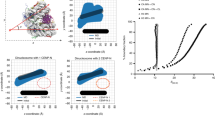
The centromere is a unique chromosomal locus that ensures accurate segregation of chromosomes during cell division by directing the assembly of a multiprotein complex, the kinetochore1. The centromere is marked by a conserved variant of conventional histone H3 termed CenH3 or CENP-A (ref. 2). A conserved motif of CenH3, the CATD, defined by loop 1 and helix 2 of the histone fold, is necessary and sufficient for specifying centromere functions of CenH3 (refs 3, 4). The structural basis of this specification is of particular interest. Yeast Scm3 and human HJURP are conserved non-histone proteins that interact physically with the (CenH3–H4)2 heterotetramer and are required for the deposition of CenH3 at centromeres in vivo5,6,7,8,9,10,11,12,13. Here we have elucidated the structural basis for recognition of budding yeast (Saccharomyces cerevisiae) CenH3 (called Cse4) by Scm3. We solved the structure of the Cse4-binding domain (CBD) of Scm3 in complex with Cse4 and H4 in a single chain model. An α-helix and an irregular loop at the conserved amino terminus and a shorter α-helix at the carboxy terminus of Scm3(CBD) wraps around the Cse4–H4 dimer. Four Cse4-specific residues in the N-terminal region of helix 2 are sufficient for specific recognition by conserved and functionally important residues in the N-terminal helix of Scm3 through formation of a hydrophobic cluster. Scm3(CBD) induces major conformational changes and sterically occludes DNA-binding sites in the structure of Cse4 and H4. These findings have implications for the assembly and architecture of the centromeric nucleosome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cleveland, D. W., Mao, Y. & Sullivan, K. F. Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell 112, 407–421 (2003)
Henikoff, S., Ahmad, K. & Malik, H. S. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293, 1098–1102 (2001)
Black, B. E. et al. Structural determinants for generating centromeric chromatin. Nature 430, 578–582 (2004)
Black, B. E. et al. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell 25, 309–322 (2007)
Mizuguchi, G., Xiao, H., Wisniewski, J., Smith, M. M. & Wu, C. Nonhistone Scm3 and histones CenH3–H4 assemble the core of centromere-specific nucleosomes. Cell 129, 1153–1164 (2007)
Camahort, R. et al. Scm3 is essential to recruit the histone H3 variant Cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell 26, 853–865 (2007)
Stoler, S. et al. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl Acad. Sci. USA 104, 10571–10576 (2007)
Camahort, R. et al. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell 35, 794–805 (2009)
Sanchez-Pulido, L., Pidoux, A. L., Pointing, C. P. & Allshire, R. C. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell 137, 1173–1174 (2009)
Williams, J. S., Hayashi, T., Yanagida, M. & Russell, P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol. Cell 33, 287–298 (2009)
Pidoux, A. L. et al. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol. Cell 33, 299–311 (2009)
Foltz, D. R. et al. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell 137, 472–484 (2009)
Dunleavy, E. M. et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497 (2009)
Malik, H. S. & Henikoff, S. Major evolutionary transitions in centromere complexity. Cell 138, 1067–1082 (2009)
Stoler, S., Keith, K. C., Curnick, K. E. & Fitzgerald-Hayes, M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9, 573–586 (1995)
Meluh, P. B., Yang, P., Glowczewski, L., Koshland, D. & Smith, M. M. Cse4p is a component of the core centromere of Saccharomyces cerevisiae . Cell 94, 607–613 (1998)
Cottarel, G., Shero, J. H., Hieter, P. & Hegemann, J. H. A 125-base-pair CEN6 DNA fragment is sufficient for complete meiotic and mitotic centromere functions in Saccharomyces cerevisiae . Mol. Cell. Biol. 9, 3342–3349 (1989)
Clarke, L. & Carbon, J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 287, 504–509 (1980)
Black, B. E. & Bassett, E. A. The histone variant CENP-A and centromere specification. Curr. Opin. Cell Biol. 20, 91–100 (2008)
Furuyama, T. & Henikoff, S. Centromeric nucleosomes induce positive DNA supercoils. Cell 138, 104–113 (2009)
Keith, K. C. et al. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol. Cell. Biol. 19, 6130–6139 (1999)
Shuaib, M., Ouararhni, K., Dimiyrov, S. & Hamiche, A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc. Natl Acad. Sci. USA 107, 1349–1354 (2010)
Wieland, G., Orthaus, S., Ohndorf, S., Diekmann, S. & Hemmerich, P. Functional complementation of human centromere protein A (CENP-A) by Cse4p from Saccharomyces cerevisiae . Mol. Cell. Biol. 24, 6620–6630 (2004)
Sekulic, N., Bassett, E. A., Rogers, D. J. & Black, B. E. The structure of (CENP-A–H4)2 reveals physical features that mark centromeres. Nature 467, 347–351 (2010)
Wood, C. M. et al. High-resolution structure of the native histone octamer. Acta Crystallogr. 61, 541–545 (2005)
English, C. M., Adkins, M. W., Carson, J. J., Churchill, M. E. & Tyler, J. K. Structural basis for the histone chaperone activity of Asf1. Cell 127, 495–508 (2006)
Natsume, R. et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature 446, 338–341 (2007)
Zhou, Z. et al. NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B. Nature Struct. Mol. Biol. 15, 868–869 (2008)
White, C. L., Suto, R. K. & Luger, K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20, 5207–5218 (2001)
Tugarinov, V., Kanelis, V. & Kay, L. E. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nature Protocols 1, 749–754 (2006)
Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity untracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 (2000)
Cole, J. L., Lary, J. W., Moody, T. P. & Laue, T. M. Analytical ultracentrifugation: sedimentation velocity and sedimentation equilibrium. Methods Cell Biol. 84, 143–179 (2008)
Schuck, P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 320, 104–124 (2003)
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995)
Johnson, B. A. & Blevins, R. A. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 (1994)
Schwieters, C. D., Kuszewski, J., Tjandra, N. & Clore, G. M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 (2003)
Zhou, Z. et al. NMR structure of chaperone Chz1 complexed with histones H2A.Z-H2B. Nature Struct. Mol. Biol. 15, 868–869 (2008)
Cornilescu, G., Delaglio, F. & Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 (1999)
Laskowski, R. A. et al. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 (1996)
Houtman, J. C. et al. Binding specificity of multiprotein signaling complexes is determined by both cooperative interactions and affinity preferences. Biochemistry 43, 4170–4178 (2004)
Acknowledgements
We thank J. Ying, K. Varney, J. F. Ellena and J. Gruschus for help collecting NMR spectra, A. Bax for discussion, C. Klee and M. Lichten for comments on the manuscript, and D. Cleveland for plasmids of human CENP-A and H4 histones. This work is supported by the intramural research programs of NCI, NIDDK and NHLBI.
Author information
Authors and Affiliations
Contributions
Z.Z. and H.F. contributed equally to this work. Z.Z. performed protein engineering, biochemical and ITC studies. B.-R.Z. contributed to protein sample preparation. B.-R.Z. and L.M.M.J. contributed to the analysis of ITC data. H.F., K.H., A.Z. and N.T. collected the NMR spectra. H.F. and Z.Z. analysed the NMR data and H.F. solved the structure. R.G. performed the sedimentation experiments. H.X. provided initial plasmids and guidance in cloning. C.W. proposed the project and participated in manuscript writing. Y.B. contributed to the overall strategy, project management and writing of the manuscript. All authors read and commented on the manuscript.
Corresponding author
Supplementary information
Supplementary Information
The file contains Supplementary Methods, Supplementary Tables 1-3, Supplementary Figures 1-21 with legends and additional references. (PDF 4692 kb)
Rights and permissions
About this article
Cite this article
Zhou, Z., Feng, H., Zhou, BR. et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature 472, 234–237 (2011). https://doi.org/10.1038/nature09854
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09854
This article is cited by
-
Uncovering natural allelic and structural variants of OsCENH3 gene by targeted resequencing and in silico mining in genus Oryza
Scientific Reports (2023)
-
Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation
Nature Communications (2021)
-
The CENP-A centromere targeting domain facilitates H4K20 monomethylation in the nucleosome by structural polymorphism
Nature Communications (2019)
-
Backbone and side-chain resonance assignments of centromeric protein Scm3 from Saccharomyces cerevisiae
Biomolecular NMR Assignments (2019)
-
Structure of centromere chromatin: from nucleosome to chromosomal architecture
Chromosoma (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.