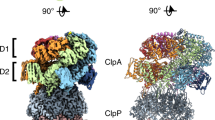
Abstract
Regulated proteolysis by ATP-dependent proteases is universal in all living cells. Bacterial ClpC, a member of the Clp/Hsp100 family of AAA+ proteins (ATPases associated with diverse cellular activities) with two nucleotide-binding domains (D1 and D2), requires the adaptor protein MecA for activation and substrate targeting. The activated, hexameric MecA–ClpC molecular machine harnesses the energy of ATP binding and hydrolysis to unfold specific substrate proteins and translocate the unfolded polypeptide to the ClpP protease for degradation. Here we report three related crystal structures: a heterodimer between MecA and the amino domain of ClpC, a heterododecamer between MecA and D2-deleted ClpC, and a hexameric complex between MecA and full-length ClpC. In conjunction with biochemical analyses, these structures reveal the organizational principles behind the hexameric MecA–ClpC complex, explain the molecular mechanisms for MecA-mediated ClpC activation and provide mechanistic insights into the function of the MecA–ClpC molecular machine. These findings have implications for related Clp/Hsp100 molecular machines.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Protein Data Bank
Data deposits
The atomic coordinates and structure factors of the ClpC N domain, the MecA121–ClpC N-domain complex, the hexameric complex of MecA121–ClpC-ΔD2 and the hexameric complex of MecA108–ClpC have been deposited in the Protein Data Bank with the accession codes 2Y1Q, 2Y1R, 3PXG and 3PXI, respectively.
References
Baker, T. A. & Sauer, R. T. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem. Sci. 31, 647–653 (2006)
Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 (2009)
Goldberg, A. L. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem. Soc. Trans. 35, 12–17 (2007)
Kirstein, J., Moliere, N., Dougan, D. A. & Turgay, K. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nature Rev. Microbiol. 7, 589–599 (2009)
Inobe, T. & Matouschek, A. Protein targeting to ATP-dependent proteases. Curr. Opin. Struct. Biol. 18, 43–51 (2008)
Groll, M., Bochtler, M., Brandstetter, H., Clausen, T. & Huber, R. Molecular machines for protein degradation. ChemBioChem 6, 222–256 (2005)
Barends, T. R., Werbeck, N. D. & Reinstein, J. Disaggregases in 4 dimensions. Curr. Opin. Struct. Biol. 20, 46–53 (2010)
Doyle, S. M. & Wickner, S. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem. Sci. 34, 40–48 (2009)
Haslberger, T., Bukau, B. & Mogk, A. Towards a unifying mechanism for ClpB/Hsp104-mediated protein disaggregation and prion propagation. Biochem. Cell Biol. 88, 63–75 (2010)
Lee, S., Sowa, M. E., Choi, J. M. & Tsai, F. T. The ClpB/Hsp104 molecular chaperone-a protein disaggregating machine. J. Struct. Biol. 146, 99–105 (2004)
Lee, S. et al. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell 115, 229–240 (2003)
Lee, S., Choi, J. M. & Tsai, F. T. Visualizing the ATPase cycle in a protein disaggregating machine: structural basis for substrate binding by ClpB. Mol. Cell 25, 261–271 (2007)
Lee, S., Sielaff, B., Lee, J. & Tsai, F. T. CryoEM structure of Hsp104 and its mechanistic implication for protein disaggregation. Proc. Natl Acad. Sci. USA 107, 8135–8140 (2010)
Wendler, P. et al. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell 131, 1366–1377 (2007)
Wendler, P. et al. Motor mechanism for protein threading through Hsp104. Mol. Cell 34, 81–92 (2009)
Wendler, P. & Saibil, H. R. Cryo electron microscopy structures of Hsp100 proteins: crowbars in or out? Biochem. Cell Biol. 88, 89–96 (2010)
Kirstein, J. et al. Adaptor protein controlled oligomerization activates the AAA+ protein ClpC. EMBO J. 25, 1481–1491 (2006)
Persuh, M., Turgay, K., Mandic-Mulec, I. & Dubnau, D. The N- and C-terminal domains of MecA recognize different partners in the competence molecular switch. Mol. Microbiol. 33, 886–894 (1999)
Turgay, K., Hamoen, L. W., Venema, G. & Dubnau, D. Biochemical characterization of a molecular switch involving the heat shock protein ClpC, which controls the activity of ComK, the competence transcription factor of Bacillus subtilis . Genes Dev. 11, 119–128 (1997)
Turgay, K., Hahn, J., Burghoorn, J. & Dubnau, D. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17, 6730–6738 (1998)
Mei, Z. et al. Molecular determinants of MecA as a degradation tag for the ClpCP protease. J. Biol. Chem. 284, 34366–34375 (2009)
Guo, F., Maurizi, M. R., Esser, L. & Xia, D. Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J. Biol. Chem. 277, 46743–46752 (2002)
Li, J. & Sha, B. Crystal structure of the E. coli Hsp100 ClpB N-terminal domain. Structure 11, 323–328 (2003)
Wang, F. et al. Crystal structure of the MecA degradation tag. J. Biol. Chem. 284, 34376–34381 (2009)
Guo, F., Esser, L., Singh, S. K., Maurizi, M. R. & Xia, D. Crystal structure of the heterodimeric complex of the adaptor, ClpS, with the N-domain of the AAA+ chaperone, ClpA. J. Biol. Chem. 277, 46753–46762 (2002)
Zeth, K. et al. Structural analysis of the adaptor protein ClpS in complex with the N-terminal domain of ClpA. Nature Struct. Biol. 9, 906–911 (2002)
Andersson, F. I. et al. Cyanobacterial ClpC/HSP100 protein displays intrinsic chaperone activity. J. Biol. Chem. 281, 5468–5475 (2006)
Lenzen, C. U., Steinmann, D., Whiteheart, S. W. & Weis, W. I. Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein. Cell 94, 525–536 (1998)
Yu, R. C., Hanson, P. I., Jahn, R. & Brunger, A. T. Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nature Struct. Biol. 5, 803–811 (1998)
Glynn, S. E., Martin, A., Nager, A. R., Baker, T. A. & Sauer, R. T. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell 139, 744–756 (2009)
Hinnerwisch, J., Fenton, W. A., Furtak, K. J., Farr, G. W. & Horwich, A. L. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell 121, 1029–1041 (2005)
Schlieker, C. et al. Substrate recognition by the AAA+ chaperone ClpB. Nature Struct. Mol. Biol. 11, 607–615 (2004)
DeLaBarre, B. & Brunger, A. T. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nature Struct. Biol. 10, 856–863 (2003)
DeLaBarre, B. & Brunger, A. T. Nucleotide dependent motion and mechanism of action of p97/VCP. J. Mol. Biol. 347, 437–452 (2005)
Davies, J. M., Brunger, A. T. & Weis, W. I. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure 16, 715–726 (2008)
Bruker . SAINT, SADABS, XPREP and SHELXTL/NT Software Reference Manual (Bruker AXS, 2003)
Kabsch, W. Evaluation of single-crystal X-ray diffraction data from a position-sensitive detector. J. Appl. Crystallogr. 21, 916–924 (1988)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative Computational Project, Number 4 . The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Schneider, T. R. & Sheldrick, G. M. Substructure solution with SHELXD. Acta Crystallogr. D 58, 1772–1779 (2002)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
DeLano, W. L. PyMOL Molecular Viewer 〈http://www.pymol.org〉 (2002)
Cook, P. F., Neville, M. E., Jr, Vrana, K. E., Hartl, F. T. & Roskoski, R., Jr Adenosine cyclic 3′,5′-monophosphate dependent protein kinase: kinetic mechanism for the bovine skeletal muscle catalytic subunit. Biochemistry 21, 5794–5799 (1982)
Weber-Ban, E. U., Reid, B. G., Miranker, A. D. & Horwich, A. L. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature 401, 90–93 (1999)
Kim, Y. I. et al. Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nature Struct. Biol. 8, 230–233 (2001)
Cowtan, K. DM: an automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newslett. Protein Crystallogr. 31, 34–38 (1994)
Cowtan, K. The Buccaneer software for automated model building. Acta Crystallogr. D 62, 1002–1011 (2006)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Acknowledgements
We thank J. He and S. Huang at Shanghai Synchrotron Radiation Source and staff at the SPring-8 beamline BL41XU for help. This work was supported by Project 30888001 of the National Natural Science Foundation of China (Y.S.) and Postdoctoral Fellowship #023201058 from the China Postdoctoral Science Foundation (Z.M.).
Author information
Authors and Affiliations
Contributions
F.W., Z.M., and Y.S. designed all experiments. F.W., Z.M., Y.Q., C.Y., Q.H., and J.W. performed the experiments. All authors analysed the data. F.W., Z.M., J.W. and Y.S. contributed to manuscript preparation. Y.S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Tables 1-2 and Supplementary Figures 1-11 with legends. (PDF 1913 kb)
Rights and permissions
About this article
Cite this article
Wang, F., Mei, Z., Qi, Y. et al. Structure and mechanism of the hexameric MecA–ClpC molecular machine. Nature 471, 331–335 (2011). https://doi.org/10.1038/nature09780
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09780
This article is cited by
-
Triple deletion of clpC, porB, and mepA enhances production of small ubiquitin-like modifier-N-terminal pro-brain natriuretic peptide in Corynebacterium glutamicum
Journal of Industrial Microbiology and Biotechnology (2019)
-
Overlapping and Specific Functions of the Hsp104 N Domain Define Its Role in Protein Disaggregation
Scientific Reports (2017)
-
An atomic structure of the human 26S proteasome
Nature Structural & Molecular Biology (2016)
-
Sending protein aggregates into a downward spiral
Nature Structural & Molecular Biology (2016)
-
Arginine phosphorylation marks proteins for degradation by a Clp protease
Nature (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.