Abstract
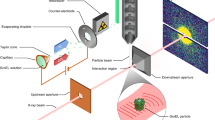
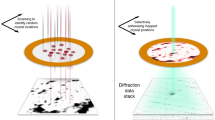
X-ray crystallography provides the vast majority of macromolecular structures, but the success of the method relies on growing crystals of sufficient size. In conventional measurements, the necessary increase in X-ray dose to record data from crystals that are too small leads to extensive damage before a diffraction signal can be recorded1,2,3. It is particularly challenging to obtain large, well-diffracting crystals of membrane proteins, for which fewer than 300 unique structures have been determined despite their importance in all living cells. Here we present a method for structure determination where single-crystal X-ray diffraction ‘snapshots’ are collected from a fully hydrated stream of nanocrystals using femtosecond pulses from a hard-X-ray free-electron laser, the Linac Coherent Light Source4. We prove this concept with nanocrystals of photosystem I, one of the largest membrane protein complexes5. More than 3,000,000 diffraction patterns were collected in this study, and a three-dimensional data set was assembled from individual photosystem I nanocrystals (∼200 nm to 2 μm in size). We mitigate the problem of radiation damage in crystallography by using pulses briefer than the timescale of most damage processes6. This offers a new approach to structure determination of macromolecules that do not yield crystals of sufficient size for studies using conventional radiation sources or are particularly sensitive to radiation damage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Owen, R. L., Rudino-Pinera, E. & Garman, E. F. Experimental determination of the radiation dose limit for cryocooled protein crystals. Proc. Natl Acad. Sci. USA 103, 4912–4917 (2006)
Henderson, R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q. Rev. Biophys. 28, 171–193 (1995)
Riekel, C. Recent developments in microdiffraction on protein crystals. J. Synchr. Radiat. 11, 4–6 (2004)
Emma, P. et al. First lasing and operation of an ångstrom-wavelength free-electron laser. Nature Photon. 4, 641–647 (2010)
Jordan, P. et al. Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411, 909–917 (2001)
Neutze, R., Wout, R., van der Spoel, D., Weckert, E. & Hajdu, J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature 406, 752–757 (2000)
Chapman, H. N. et al. Femtosecond time-delay X-ray holography. Nature 448, 676–679 (2007)
Spence, J. C. H. & Doak, R. B. Single molecule diffraction. Phys. Rev. Lett. 92, 198102 (2004)
DePonte, D. P. et al. Gas dynamic virtual nozzle for generation of microscopic droplet streams. J. Phys. D 41, 195505 (2008)
Henderson, R. et al. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J. Mol. Biol. 213, 899–929 (1990)
Wang, D. N. & Kühlbrandt, W. High-resolution electron crystallography of light-harvesting chlorophyll a/b-protein complex in three different media. J. Mol. Biol. 217, 691–699 (1991)
Strüder, L. et al. Large-format, high-speed, X-ray pnCCDs combined with electron and ion imaging spectrometers in a multipurpose chamber for experiments at 4th generation light sources. Nucl. Instrum. Methods Phys. Res. A 614, 483–496 (2010)
Ding, Y. et al. Measurements and simulations of ultralow emittance and ultrashort electron beams in the Linac Coherent Light Source. Phys. Rev. Lett. 102, 254801 (2009)
Paithankar, K. S., Owen, R. L. & Garman, E. F. Absorbed dose calculations for macromolecular crystals: improvements to RADDOSE. J. Synchr. Radiat. 16, 152–162 (2009)
Marchesini, S. et al. X-ray image reconstruction from a diffraction pattern alone. Phys. Rev. B 68, 140101 (2003)
Robinson, I. K. & Harder, R. Coherent X-ray diffraction imaging of strain at the nanoscale. Nature Mater. 8, 291–298 (2009)
Sayre, D. Some implications of a theorem due to Shannon. Acta Crystallogr. 5, 843 (1952)
Kirian, R. et al. Femtosecond protein nanocrystallography—data analysis methods. Opt. Express 18, 5713–5723 (2010)
Leslie, A. G. The integration of macromolecular diffraction data. Acta Crystallogr. D 62, 48–57 (2006)
Duisenberg, A. J. M. Indexing in single-crystal diffractometry with an obstinate list of reflections. J. Appl. Cryst. 25, 92–96 (1992)
Young, L. et al. Femtosecond electronic response of atoms to ultra-intense X-rays. Nature 466, 56–61 (2010)
Hau-Riege, S. P., London, R. A. & Szoke, A. Dynamics of biological molecules irradiated by short X-ray pulses. Phys. Rev. E 69, 051906 (2004)
Bergh, M., Huldt, G., Timneanu, N., Maia, F. R. N. C. & Hajdu, J. Feasibility of imaging living cells at subnanometer resolutions by ultrafast X-ray diffraction. Q. Rev. Biophys. 41, 181–204 (2008)
Willis, B. & Pryor, A. Thermal Vibrations in Crystallography 92 (Cambridge Univ. Press, 1975)
Emma, P. et al. Femtosecond and subfemtosecond X-ray pulses from a self-amplified spontaneous-emission based free-electron laser. Phys. Rev. Lett. 92, 074801 (2004)
Loh, N.-T. D. & Elser, V. Reconstruction algorithm for single-particle diffraction imaging experiments. Phys. Rev. E 80, 026705 (2009)
Fung, R., Shneerson, V., Saldin, D. K. & Ourmazd, A. Structure from fleeting illumination of faint spinning objects in flight. Nature Phys. 5, 64–67 (2008)
Rossmann, M. G., Leslie, A. G., Sherin, S. A. & Tsukihara, T. Processing and post-refinement of oscillation camera data. J. Appl. Cryst. 12, 570–581 (1979)
Bozek, J. D. AMO instrumentation for the LCLS X-ray FEL. Eur. Phys. J. Spec. Top. 169, 129–132 (2009)
DePonte, D. P. et al. SEM imaging of liquid jets. Micron 40, 507–509 (2009)
Fromme, P. & Grotjohann, I. in Membrane Protein Crystallization (ed. DeLukas, L. ) 192–224 (Curr. Top. Membr. 63, Elsevier, 2009)
Hunter, M. S. et al. X-ray diffraction from membrane protein nanocrystals. Biophys. J. (in the press)
Zaefferer, S. New developments of computer-aided crystallographic analysis in transmission electron microscopy. J. Appl. Cryst. 33, 10–25 (2000)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likehood method. Acta Crystallogr. D 53, 240–255 (1997)
Praznikar, J., Afonine, P. V., Guncar, G., Adams, P. D. & Turk, D. Averaged kick maps: less noise, more signal…and probably less bias. Acta Crystallogr. D 65, 921–931 (2009)
Acknowledgements
Experiments were carried out at the Linac Coherent Light Source and the Advanced Light Source, both National User Facilities operated respectively by Stanford University and the University of California on behalf of the US Department of Energy (DOE), Office of Basic Energy Sciences. We acknowledge support from the DOE through the PULSE Institute at the SLAC National Accelerator Laboratory; the Lawrence Livermore National Laboratory under contract DE-AC52-07NA27344; the Center for Bio-Inspired Solar Fuel Production, an Energy Frontier Research Center funded by the DOE, Office of Basic Energy Sciences (award DE-SC0001016); the Hamburg Ministry of Science and Research and the Joachim Herz Stiftung, as part of the Hamburg Initiative for Excellence in Research (LEXI); the Hamburg School for Structure and Dynamics; the Max Planck Society, for funding the development and operation of the CAMP instrument within the ASG at CFEL; the US National Science Foundation (awards 0417142 and MCB-1021557); the US National Institutes of Health (awards 1R01GM095583-01 (ROADMAP) and 1U54GM094625-01 (PSI:Biology)); the Swedish Research Council; the Swedish Foundation for International Cooperation in Research and Higher Education; Stiftelsen Olle Engkvist Byggmästare; the DFG Cluster of Excellence at the Munich Centre for Advanced Photonics; and the CBST at the University of California under cooperative agreement no. PHY 0120999. We acknowledge discussions with M. Rossmann, E. Snell, R. Stroud and A. Brunger, thank B. Hedman, E. Gullikson, F. Filsinger, A. Berg, H. Mahn and C. Kaiser for technical help and thank the staff of the LCLS for their support in carrying out these experiments.
Author information
Authors and Affiliations
Contributions
H.N.C. and J.C.H.S. conceived the experiment, which was designed with P.F., A.B., R.A.K., J.S., D.P.D., U.W., R.B.D., S. Boutet, M.J.B., D.S., I.S., S.M. and J.H. The CAMP instrument was the responsibility of S.W.E., R.H., D. Rolles, A. Rudenko, C.S., L.F., N.K., P.H., B.R., B.E., A.H., Ch.R., D.P., G.W., L.S., G.H., H. Gorke, J.U., I.S., S.H., G.S., F.S., H.S., K.-U.K., R.A., C.-D.S., F.K., M. Bott, S. Schorb, D. Rupp, M.A., T.G., H.H., L.G., G.P., H. Graafsma and B.N., who designed and set up the instrument and/or developed and operated the pnCCD detectors. C.B., J.D.B. and M.M. set up and aligned the beamline. P.F., M.S.H. and I.G. prepared samples; R.B.D., D.P.D., U.W., J.C.H.S., P.F., L.L. and R.L.S. developed and operated the sample delivery system; H.N.C., A.B., A.A., J.S., D.P.D., U.W., R.B.D., S. Bajt, M.J.B., L.G., J.H., M.M.S., N.T., J.A., S. Stern and J.C.H.S. developed diffraction instrumentation; and M. Barthelmess, M.L., A.B. and K.N. designed and/or fabricated calibration samples. J.K., S.P.H.-R., A.B., H.N.C., J.S. and A.V.M. characterized the focus. H.N.C., J.C.H.S., P.F., A.B., T.A.W., R.A.K., A.A., J.S., D.P.D., U.W., R.B.D., I.S., N.C., R.L.S., M.S.H., L.L., M. Bott, S.W.E., R.H., D. Rolles, A. Rudenko, M.L., C.B., J.U., L.F., J.D.B., M.M., M.F., C.Y.H., R.G.S., G.J.W., A. Rocker, M.S., O.J., I.A. and J.H. carried out the experiment. A.B., T.A.W., R.A.K., A.A., F.R.N.C.M., A.V.M., L.L., T.R.M.B., N.C., L.F., N.K., R.N., G.W., P.H., C.C., J.M.H., I.S., J.H., H.N.C. and J.C.H.S. analysed the data. A.V.M. performed the Bragg shape phase retrieval. T.A.W. and R.A.K. merged the 3D data. R.F. collected and evaluated the reference data set; R.A.K., T.A.W., J.M.H. and R.F. refined the structure and calculated the electron density maps; and H.N.C., P.F., J.C.H.S. and I.S. wrote the manuscript with discussion and improvements from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Table 1, Supplementary Figures 1-4 with legends, Supplementary Methods and Data and additional references. (PDF 9289 kb)
Rights and permissions
About this article
Cite this article
Chapman, H., Fromme, P., Barty, A. et al. Femtosecond X-ray protein nanocrystallography. Nature 470, 73–77 (2011). https://doi.org/10.1038/nature09750
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09750
This article is cited by
-
3D atomic structure from a single X-ray free electron laser pulse
Nature Communications (2024)
-
Dynamic three-dimensional structures of a metal–organic framework captured with femtosecond serial crystallography
Nature Chemistry (2024)
-
Chaotic dynamics in X-ray free-electron lasers with an optical undulator
Scientific Reports (2024)
-
Attosecond electron microscopy by free-electron homodyne detection
Nature Photonics (2024)
-
A guideline for the distance measurement plans of site-directed spin labels for structural prediction of nucleic acids
Journal of Molecular Modeling (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.