Abstract
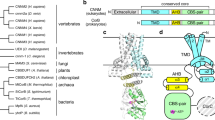
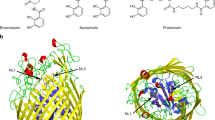
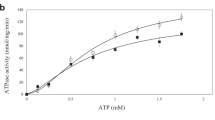
The energy-coupling factor (ECF) transporters, responsible for vitamin uptake in prokaryotes, are a unique family of membrane transporters1,2. Each ECF transporter contains a membrane-embedded, substrate-binding protein (known as the S component), an energy-coupling module that comprises two ATP-binding proteins (known as the A and A′ components) and a transmembrane protein (known as the T component). The structure and transport mechanism of the ECF family remain unknown. Here we report the crystal structure of RibU, the S component of the ECF-type riboflavin transporter from Staphylococcus aureus at 3.6-Å resolution. RibU contains six transmembrane segments, adopts a previously unreported transporter fold and contains a riboflavin molecule bound to the L1 loop and the periplasmic portion of transmembrane segments 4–6. Structural analysis reveals the essential ligand-binding residues, identifies the putative transport path and, with sequence alignment, uncovers conserved structural features and suggests potential mechanisms of action among the ECF transporters.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Rodionov, D. A. et al. A novel class of modular transporters for vitamins in prokaryotes. J. Bacteriol. 191, 42–51 (2009)
Rodionov, D. A., Hebbeln, P., Gelfand, M. S. & Eitinger, T. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 188, 317–327 (2006)
Hollenstein, K., Dawson, R. J. & Locher, K. P. Structure and mechanism of ABC transporter proteins. Curr. Opin. Struct. Biol. 17, 412–418 (2007)
Rees, D. C., Johnson, E. & Lewinson, O. ABC transporters: the power to change. Nature Rev. Mol. Cell Biol. 10, 218–227 (2009)
Davidson, A. L., Dassa, E., Orelle, C. & Chen, J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72, 317–364 (2008)
Oldham, M. L., Davidson, A. L. & Chen, J. Structural insights into ABC transporter mechanism. Curr. Opin. Struct. Biol. 18, 726–733 (2008)
Linton, K. J. Structure and function of ABC transporters. Physiology 22, 122–130 (2007)
Dawson, R. J., Hollenstein, K. & Locher, K. P. Uptake or extrusion: crystal structures of full ABC transporters suggest a common mechanism. Mol. Microbiol. 65, 250–257 (2007)
Davidson, A. L. & Maloney, P. C. ABC transporters: how small machines do a big job. Trends Microbiol. 15, 448–455 (2007)
Locher, K. P. Review. Structure and mechanism of ATP-binding cassette transporters. Phil. Trans. R. Soc. B 364, 239–245 (2009)
Jones, P. M., O’Mara, M. L. & George, A. M. ABC transporters: a riddle wrapped in a mystery inside an enigma. Trends Biochem. Sci. 34, 520–531 (2009)
Hebbeln, P., Rodionov, D. A., Alfandega, A. & Eitinger, T. Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc. Natl Acad. Sci. USA 104, 2909–2914 (2007)
Burgess, C. M. et al. The riboflavin transporter RibU in Lactococcus lactis: molecular characterization of gene expression and the transport mechanism. J. Bacteriol. 188, 2752–2760 (2006)
Duurkens, R. H., Tol, M. B., Geertsma, E. R., Permentier, H. P. & Slotboom, D. J. Flavin binding to the high affinity riboflavin transporter RibU. J. Biol. Chem. 282, 10380–10386 (2007)
Vogl, C. et al. Characterization of riboflavin (vitamin B2) transport proteins from Bacillus subtilis and Corynebacterium glutamicum . J. Bacteriol. 189, 7367–7375 (2007)
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 (1993)
Lieberman, R. L. & Rosenzweig, A. C. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature 434, 177–182 (2005)
Hakemian, A. S. et al. The metal centers of particulate methane monooxygenase from Methylosinus trichosporium OB3b. Biochemistry 47, 6793–6801 (2008)
Neubauer, O. et al. Two essential arginine residues in the T components of energy-coupling factor transporters. J. Bacteriol. 191, 6482–6488 (2009)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
DeLano, W. L. The PyMOL molecular graphics system. <http://www.pymol.org> (2002)
Bandrin, S. V., Rabinovich, P. M. & Stepanov, A. I. Three linkage groups of the genes of riboflavin biosynthesis in Escherichia coli . Genetika 19, 1419–1925 (1983)
Acknowledgements
We thank H. Yan for technical advice, A. Schmedel for administrative assistance and E. coli Genetic Resources at Yale Coli Genetic Stock Center for providing mutant E. coli strains. This work was supported by the National Institutes of Health (R01 GM084964), funds from the Ministry of Science and Technology of China (grant number 2009CB918801) and Project 30888001 supported by National Natural Science Foundation of China.
Author information
Authors and Affiliations
Contributions
P.Z. and Y.S. designed all experiments. P.Z. performed the bulk of the experiments. P.Z., J.W. and Y.S. analysed the data and contributed to manuscript preparation. Y.S. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Table 1 and Supplementary Figures 1-15 with legends. (PDF 2565 kb)
Rights and permissions
About this article
Cite this article
Zhang, P., Wang, J. & Shi, Y. Structure and mechanism of the S component of a bacterial ECF transporter. Nature 468, 717–720 (2010). https://doi.org/10.1038/nature09488
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09488
This article is cited by
-
Conserved and lineage-specific hypothetical proteins may have played a central role in the rise and diversification of major archaeal groups
BMC Biology (2022)
-
Membrane mediated toppling mechanism of the folate energy coupling factor transporter
Nature Communications (2020)
-
Cu Transport by the Extended Family of CcoA-like Transporters (CalT) in Proteobacteria
Scientific Reports (2019)
-
Structure and mechanism of a group-I cobalt energy coupling factor transporter
Cell Research (2017)
-
Structural insight in the toppling mechanism of an energy-coupling factor transporter
Nature Communications (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.