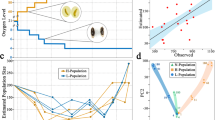
Abstract
Experimental evolution systems allow the genomic study of adaptation, and so far this has been done primarily in asexual systems with small genomes, such as bacteria and yeast1,2,3. Here we present whole-genome resequencing data from Drosophila melanogaster populations that have experienced over 600 generations of laboratory selection for accelerated development. Flies in these selected populations develop from egg to adult ∼20% faster than flies of ancestral control populations, and have evolved a number of other correlated phenotypes. On the basis of 688,520 intermediate-frequency, high-quality single nucleotide polymorphisms, we identify several dozen genomic regions that show strong allele frequency differentiation between a pooled sample of five replicate populations selected for accelerated development and pooled controls. On the basis of resequencing data from a single replicate population with accelerated development, as well as single nucleotide polymorphism data from individual flies from each replicate population, we infer little allele frequency differentiation between replicate populations within a selection treatment. Signatures of selection are qualitatively different than what has been observed in asexual species; in our sexual populations, adaptation is not associated with ‘classic’ sweeps whereby newly arising, unconditionally advantageous mutations become fixed. More parsimonious explanations include ‘incomplete’ sweep models, in which mutations have not had enough time to fix, and ‘soft’ sweep models, in which selection acts on pre-existing, common genetic variants. We conclude that, at least for life history characters such as development time, unconditionally advantageous alleles rarely arise, are associated with small net fitness gains or cannot fix because selection coefficients change over time.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
GenBank/EMBL/DDBJ
Data deposits
The FASTQ files associated with this project have been deposited in GenBank's Short Read Archive under the study accession number SRP002024. Data and source code files to reproduce the analyses of this work are available on request from the authors.
References
Papadopoulos, D. et al. Genomic evolution during a 10,000-generation experiment with bacteria. Proc. Natl Acad. Sci. USA 96, 3807–3812 (1999)
Dunham, M. J. et al. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae . Proc. Natl Acad. Sci. USA 99, 16144–16149 (2002)
Hegreness, M. & Kishony, R. Analysis of genetic systems using experimental evolution and whole-genome sequencing. Genome Biol. 8, 201 (2007)
Garland, T. & Rose, M. R. Experimental Evolution: Concepts, Methods, and Applications of Selection Experiments (Univ. California Press, 2009)
Teotónio, H., Chelo, I. M., Bradic, M., Rose, M. R. & Long, A. D. Experimental evolution reveals natural selection on standing genetic variation. Nature Genet. 41, 251–257 (2009)
Herring, C. D. et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nature Genet. 38, 1406–1412 (2006)
Barrick, J. E. et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli . Nature 461, 1243–1247 (2009)
Hermisson, J. & Pennings, P. S. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics 169, 2335–2352 (2005)
Feder, J. L. et al. Allopatric genetic origins for sympatric host-plant shifts and race formation in Rhagoletis. Proc. Natl Acad. Sci. USA 100, 10314–10319 (2003)
Pelz, H. J. et al. The genetic basis of resistance to anticoagulants in rodents. Genetics 170, 1839–1847 (2005)
Colosimo, P. F. et al. Widespread parallel evolution in sticklebacks by repeated fixation of ectodysplasin alleles. Science 307, 1928–1933 (2005)
Przeworski, M., Coop, G. & Wall, J. D. The signature of positive selection on standing genetic variation. Evolution 59, 2312–2323 (2005)
Rose, M. R., Passananti, H. B. & Matos, M. Methuselah Flies: A Case Study in the Evolution of Aging (World Scientific, 2004)
Hoekstra, H. E. & Coyne, J. A. The locus of evolution: evo devo and the genetics of adaptation. Evolution 61, 995–1016 (2007)
Dennis, G. et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 4, R60 (2003)
Kaplan, N. L., Hudson, R. R. & Langley, C. H. The hitchhiking effect revisited. Genetics 123, 887–899 (1989)
Innan, H. & Kim, Y. Pattern of polymorphism after strong artificial selection in a domestication event. Proc. Natl Acad. Sci. USA 101, 10667–10672 (2004)
Teotónio, H. & Rose, M. R. Variation in the reversibility of evolution. Nature 408, 463–466 (2000)
Chevin, L. M. & Hospital, F. Selective sweep at a quantitative trait locus in the presence of background genetic variation. Genetics 180, 1645–1660 (2008)
Chippindale, A. K., Alipaz, J. A., Chen, H. W. & Rose, M. R. Experimental evolution of accelerated development in Drosophila. 1. Developmental speed and larval survival. Evolution 51, 1536–1551 (1997)
Chippindale, A. K., Alipaz, J. A. & Rose, M. R. in Methuselah Flies (eds Rose, M. R., Passananti, H. B. & Matos, M.) 413–435 (World Scientific, 2004)
Bloom, J. S., Khan, Z., Kruglyak, L., Singh, M. & Caudy, A. A. Measuring differential gene expression by short read sequencing: quantitative comparison to 2-channel gene expression microarrays. BMC Genomics 10, 221 (2009)
Acknowledgements
We thank K. Aeling and D. Heck at the UCI DNA and Protein Microarray Facility for help with preparation and sequencing of the ACO1 library. The pooled ACO and CO libraries were sequenced at the University of Oregon High-Throughput Sequencing Facility, with advice from D. Turnbull. We are grateful to S. Nuzdhin, T. Turner and J. J. Emerson for providing suggestions during the conception of the project, and to O. Tenaillon and F. Barreto for comments on previous versions of the manuscript. We also thank A. K. Chippindale for discussion of the phenotype data. This work was supported by a UCI Faculty Research and Training Grant to M.R.R. and NSF DEB-0614429 to A.D.L. M.K.B. is supported by an NSF Graduate Fellowship in STEM K-12 Education (DGE-0638751).
Author information
Authors and Affiliations
Contributions
M.K.B., P.S. and J.P.D. performed the laboratory experiments. M.K.B., K.R.T. and A.D.L. analysed the data. M.K.B., M.R.R. and A.D.L. designed the project, and M.K.B., K.R.T., M.R.R. and A.D.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-7 with legends and Supplementary Tables 1-2. (PDF 1877 kb)
Rights and permissions
About this article
Cite this article
Burke, M., Dunham, J., Shahrestani, P. et al. Genome-wide analysis of a long-term evolution experiment with Drosophila. Nature 467, 587–590 (2010). https://doi.org/10.1038/nature09352
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09352
This article is cited by
-
Haplotype based testing for a better understanding of the selective architecture
BMC Bioinformatics (2023)
-
Revisiting the Design of the Long-Term Evolution Experiment with Escherichia coli
Journal of Molecular Evolution (2023)
-
Embracing Complexity: Yeast Evolution Experiments Featuring Standing Genetic Variation
Journal of Molecular Evolution (2023)
-
Identifying Targets of Selection in Laboratory Evolution Experiments
Journal of Molecular Evolution (2023)
-
Uncovering the mode of action of engineered T cells in patient cancer organoids
Nature Biotechnology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.