Abstract
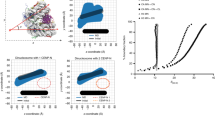
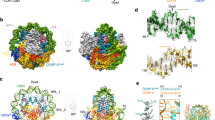
Centromeres are specified epigenetically, and the histone H3 variant CENP-A is assembled into the chromatin of all active centromeres1. Divergence from H3 raises the possibility that CENP-A generates unique chromatin features to mark physically centromere location. Here we report the crystal structure of a subnucleosomal heterotetramer, human (CENP-A–H4)2, that reveals three distinguishing properties encoded by the residues that comprise the CENP-A targeting domain (CATD; ref. 2): (1) a CENP-A–CENP-A interface that is substantially rotated relative to the H3–H3 interface; (2) a protruding loop L1 of the opposite charge as that on H3; and (3) strong hydrophobic contacts that rigidify the CENP-A–H4 interface. Residues involved in the CENP-A–CENP-A rotation are required for efficient incorporation into centromeric chromatin, indicating specificity for an unconventional nucleosome shape. DNA topological analysis indicates that CENP-A-containing nucleosomes are octameric with conventional left-handed DNA wrapping, in contrast to other recent proposals3,4,5,6. Our results indicate that CENP-A marks centromere location by restructuring the nucleosome from within its folded histone core.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Black, B. E. & Bassett, E. A. The histone variant CENP-A and centromere specification. Curr. Opin. Cell Biol. 20, 91–100 (2008)
Black, B. E. et al. Structural determinants for generating centromeric chromatin. Nature 430, 578–582 (2004)
Mizuguchi, G., Xiao, H., Wisniewski, J., Smith, M. M. & Wu, C. Nonhistone Scm3 and histones CenH3–H4 assemble the core of centromere-specific nucleosomes. Cell 129, 1153–1164 (2007)
Williams, J. S., Hayashi, T., Yanagida, M. & Russell, P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric chromatin. Mol. Cell 33, 287–298 (2009)
Dalal, Y., Wang, H., Lindsay, S. & Henikoff, S. Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol. 5, e218 (2007)
Furuyama, T. & Henikoff, S. Centromeric nucleosomes induce positive DNA supercoils. Cell 138, 104–113 (2009)
Smith, M. M. Centromeres and variant histones: what, where, when and why? Curr. Opin. Cell Biol. 14, 279–285 (2002)
Henikoff, S., Ahmad, K. & Malik, H. S. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293, 1098–1102 (2001)
Black, B. E. et al. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol. Cell 25, 309–322 (2007)
Erhardt, S. et al. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J. Cell Biol. 183, 805–818 (2008)
Shelby, R. D., Vafa, O. & Sullivan, K. F. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 136, 501–513 (1997)
Camahort, R. et al. Cse4 is part of an octameric nucleosome in budding yeast. Mol. Cell 35, 794–805 (2009)
Black, B. E., Brock, M. A., Bédard, S., Woods, V. L. & Cleveland, D. W. An epigenetic mark generated by the incorporation of CENP-A into centromeric nucleosomes. Proc. Natl Acad. Sci. USA 104, 5008–5013 (2007)
Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997)
Davey, C. A., Sargent, D. F., Luger, K., Maeder, A. W. & Richmond, T. J. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 319, 1097–1113 (2002)
Altaf, M. et al. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol. Cell 28, 1002–1014 (2007)
Lu, X. et al. The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nature Struct. Mol. Biol. 15, 1122–1124 (2008)
Englander, S. W. Hydrogen exchange and mass spectrometry: a historical perspective. J. Am. Soc. Mass Spectrom. 17, 1481–1489 (2006)
Lusser, A. & Kadonaga, J. T. Strategies for the reconstitution of chromatin. Nature Methods 1, 19–26 (2004)
Carruthers, L. M., Tse, C., Walker, K. P. & Hansen, J. C. Assembly of defined nucleosomal and chromatin arrays from pure components. Methods Enzymol. 304, 19–35 (1999)
Conde e Silva, N. et al. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J. Mol. Biol. 370, 555–573 (2007)
Simpson, R. T., Thoma, F. & Brubaker, J. M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell 42, 799–808 (1985)
Esposito, F. & Sinden, R. R. Supercoiling in prokaryotic and eukaryotic DNA: changes in response to topological perturbation of plasmids in E. coli and SV40 in vitro, in nuclei and in CV-1 cells. Nucleic Acids Res. 15, 5105–5124 (1987)
Shuaib, M., Ouararhni, K., Dimitrov, S. & Hamiche, A. HJURP binds CENP-A via a highly conserved N-terminal domain and mediates its deposition at centromeres. Proc. Natl Acad. Sci. USA 107, 1349–1354 (2010)
Bina-Stein, M. & Simpson, R. T. Specific folding and contraction of DNA by histones H3 and H4. Cell 11, 609–618 (1977)
Bassett, E. A. et al. Epigenetic centromere specification directs Aurora B accumulation but is insufficient to efficiently correct mitotic errors. J. Cell Biol. 190, 177–185 (2010)
Vermaak, D., Hayden, H. S. & Henikoff, S. Centromere targeting element within the histone fold domain of Cid. Mol. Cell. Biol. 22, 7553–7561 (2002)
Foltz, D. R. et al. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell 137, 472–484 (2009)
Carroll, C. W., Silva, M. C. C., Godek, K. M., Jansen, L. E. T. & Straight, A. F. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nature Cell Biol. 11, 896–902 (2009)
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 (1993)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Dodson, E. J., Winn, M. & Ralph, A. Collaborative Computational Project, number 4: providing programs for protein crystallography. Methods Enzymol. 277, 620–633 (1997)
Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nature Protocols 3, 1171–1179 (2008)
Konarev, P. V., Volkov, V. V., Sokolova, A. V., Koch, M. H. & Svergun, D. I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 (2003)
Svergun, D. I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 (1992)
Svergun, D., Barberato, C. & Koch, M. H. J. CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 (1995)
Wriggers, W. Using Situs for the integration of multi-resolution structures. Biophys. Rev. 2, 21–27 (2010)
Volkov, V. V. & Svergun, D. I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 36, 860–864 (2003)
Pettersen, E. F. et al. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Fyodorov, D. V. & Kadonaga, J. T. Chromatin assembly in vitro with purified recombinant ACF and NAP-1. Methods Enzymol. 371, 499–515 (2003)
Acknowledgements
We thank D. Cleveland for plasmids and steadfast encouragement to pursue a physical understanding of the centromere; S. Wood for generating cleaved histone H2A; K. Gupta for help with collecting data and technical suggestions; and K. Ferguson, G. Van Duyne, M. Lemmon, J. Shorter, L. Jansen, D. Foltz, J. Shah, D. Alvarado, K. Moravcevic and T. Panchenko for discussions and comments on the manuscript. This work was supported by the NIH research grant GM82989, a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund, and a Rita Allen Foundation Scholar Award to B.E.B. N.S. is supported by a postdoctoral fellowship from the American Cancer Society and E.A.B. has been supported by the Penn Structural Biology Training Grant (NIH GM08275) and a predoctoral fellowship from the American Heart Association.
Author information
Authors and Affiliations
Contributions
N.S. designed and performed experiments, solved and refined the structures, analysed data and wrote the manuscript; E.A.B. and D.J.R. performed experiments and analysed data; and B.E.B. directed the project, designed and performed experiments, analysed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Table 1, Supplementary Figures 1-12 with legends and additional References. (PDF 5830 kb)
Rights and permissions
About this article
Cite this article
Sekulic, N., Bassett, E., Rogers, D. et al. The structure of (CENP-A–H4)2 reveals physical features that mark centromeres. Nature 467, 347–351 (2010). https://doi.org/10.1038/nature09323
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09323
This article is cited by
-
CENP-A and CENP-B collaborate to create an open centromeric chromatin state
Nature Communications (2023)
-
Characterization of centromeric DNA of Gossypium anomalum reveals sequence-independent enrichment dynamics of centromeric repeats
Chromosome Research (2023)
-
Histone variant H2A.B-H2B dimers are spontaneously exchanged with canonical H2A-H2B in the nucleosome
Communications Biology (2021)
-
Structural alteration of DNA induced by viral protein R of HIV-1 triggers the DNA damage response
Retrovirology (2018)
-
Posttranslational modifications of CENP-A: marks of distinction
Chromosoma (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.