Abstract
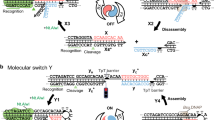
Our ability to synthesize nanometre-scale chemical species, such as nanoparticles with desired shapes and compositions, offers the exciting prospect of generating new functional materials and devices by combining them in a controlled fashion into larger structures. Self-assembly can achieve this task efficiently, but may be subject to thermodynamic and kinetic limitations: reactants, intermediates and products may collide with each other throughout the assembly time course to produce non-target species instead of target species. An alternative approach to nanoscale assembly uses information-containing molecules such as DNA1 to control interactions and thereby minimize unwanted cross-talk between different components. In principle, this method should allow the stepwise and programmed construction of target products by linking individually selected nanoscale components—much as an automobile is built on an assembly line. Here we demonstrate that a nanoscale assembly line can be realized by the judicious combination of three known DNA-based modules: a DNA origami2 tile that provides a framework and track for the assembly process, cassettes containing three independently controlled two-state DNA machines that serve as programmable cargo-donating devices3,4 and are attached4,5 in series to the tile, and a DNA walker that can move on the track from device to device and collect cargo. As the walker traverses the pathway prescribed by the origami tile track, it sequentially encounters the three DNA devices, each of which can be independently switched between an ‘ON’ state, allowing its cargo to be transferred to the walker, and an ‘OFF’ state, in which no transfer occurs. We use three different types of gold nanoparticle species as cargo and show that the experimental system does indeed allow the controlled fabrication of the eight different products that can be obtained with three two-state devices.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Seeman, N. C. & Lukeman, P. S. Nucleic acid nanostructures. Rep. Prog. Phys. 68, 237–270 (2005)
Rothemund, P. W. K. Scaffolded DNA origami for nanoscale shapes and patterns. Nature 440, 297–302 (2006)
Yan, H., Zhang, X., Shen, Z. & Seeman, N. C. A robust DNA mechanical device controlled by hybridization topology. Nature 415, 62–65 (2002)
Ding, B. & Seeman, N. C. Operation of a DNA robot arm inserted into a 2D DNA crystalline substrate. Science 314, 1583–1585 (2006)
Gu, H., Chao, J., Xiao, S. J. & Seeman, N. C. Dynamic patterning programmed by DNA tiles captured on a DNA origami substrate. Nature Nanotechnol. 4, 245–249 (2009)
Ding, B., Sha, R. & Seeman, N. C. Pseudohexagonal 2D DNA crystals from double crossover cohesion. J. Am. Chem. Soc. 126, 10230–10231 (2004)
Constantinou, P. E. et al. Double cohesion in structural DNA nanotechnology. Org. Biomol. Chem. 4, 3414–3419 (2006)
Sherman, W. B. & Seeman, N. C. A precisely controlled DNA bipedal walking device. Nano Lett. 4, 1203–1207 (2004)
Shin, J. S. & Pierce, N. A. A synthetic DNA walker for molecular transport. J. Am. Chem. Soc. 126, 10834–10835 (2004)
Bath, J., Green, S. J., Allen, K. E. & Turberfield, A. J. Mechanism for a directional, processive and reversible DNA walker. Small 5, 1513–1516 (2009)
Omabegho, T., Sha, R. & Seeman, N. C. A bipedal DNA Brownian motor with coordinated legs. Science 324, 67–71 (2009)
Liu, D., Wang, W., Deng, Z., Walulu, R. & Mao, C. Tensegrity: construction of rigid DNA triangles with flexible four-arm junctions. J. Am. Chem. Soc. 126, 2324–2325 (2004)
Yurke, B., Turberfield, A. J., Mills, A. P. Jr, Simmel, F. C. & Newmann, J. L. A DNA-fuelled molecular machine made of DNA. Nature 406, 605–608 (2000)
Drexler, K. E. Machine-phase nanotechnology. Sci. Am. 285, 74–75 (2001)
Smalley, R. E. Of chemistry, love and nanobots. Sci. Am. 285, 76–77 (2001)
Gartner, Z. J., Kanan, M. W. & Liu, D. R. Multi-step small molecule synthesis programmed by DNA templates. J. Am. Chem. Soc. 124, 10304–10306 (2002)
Seeman, N. C. De novo design of sequences for nucleic acid structure engineering. J. Biomol. Struct. Dyn. 8, 573–581 (1990)
Caruthers, M. H. Gene synthesis machines: DNA chemistry and its uses. Science 230, 281–285 (1985)
Ke, Y., Lindsay, S., Chang, Y., Liu, Y. & Yan, H. Self-assembled water-soluble nucleic acid probe tiles for label-free RNA hybridization assays. Science 319, 180–183 (2008)
Zheng, J. et al. Two-dimensional nanoparticle arrays show the organizational power of robust DNA motifs. Nano Lett. 6, 1502–1504 (2006)
Alivisatos, A. P. et al. Organization of ‘nanocrystal molecules’ using DNA. Nature 382, 609–611 (1996)
Acknowledgements
We are grateful to J. Canary, H. Yan, C. Mao and R. Sha for comments on this manuscript. This research has been supported by the following grants: GM-29544 from the US National Institute of General Medical Sciences, CTS-0608889 and CCF-0726378 from the US National Science Foundation, 48681-EL and W911NF-07-1-0439 from the US Army Research Office, N000140910181 and N000140911118 from the US Office of Naval Research and a grant from the W. M. Keck Foundation (to N.C.S.); and 2007CB925101 from the National Basic Research Program of China and 20721002 from the National Natural Science Foundation of China (to S.-J.X.). H.G. thanks New York University for a Dissertation Fellowship and J.C. thanks the Chinese Scholarship Council for a research fellowship.
Author information
Authors and Affiliations
Contributions
H.G. and J.C. did the research, analysed data and wrote the paper. S.-J.X. analysed data and wrote the paper. N.C.S. designed the project, analysed data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file comprises Supplementary Figures S1- S12 with legends and Supplementary Table S1 as follows: origami design (figure S1) and sequence; cassette design and sequence (figures S2-S4); walker locomotion and sequence design (figure S5); nondenaturing gels of the cassettes and walker (S6); agarose gels of the DNA-nanoparticle conjugates (figure S7), AFM images of the movement of the walker on the origami (figure S8); AFM images of the dynamic switching of each cassette on the origami (figure S9); zoomed TEM images for statistical analysis (figures S10 and S11); the distance distribution between particles on the walker (figure S12), and the statistical analysis for the different additions (table S1). (PDF 4267 kb)
Rights and permissions
About this article
Cite this article
Gu, H., Chao, J., Xiao, SJ. et al. A proximity-based programmable DNA nanoscale assembly line. Nature 465, 202–205 (2010). https://doi.org/10.1038/nature09026
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature09026
This article is cited by
-
Label-free detection of biomolecules using inductively coupled plasma mass spectrometry (ICP-MS)
Analytical and Bioanalytical Chemistry (2024)
-
Computational analysis of protein synthesis, diffusion, and binding in compartmental biochips
Microbial Cell Factories (2023)
-
Adsorbate motors for unidirectional translation and transport
Nature (2023)
-
DNA double helix, a tiny electromotor
Nature Nanotechnology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.