Abstract
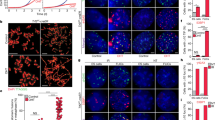
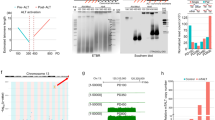
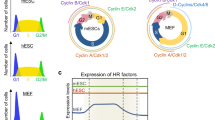
Exceptional genomic stability is one of the hallmarks of mouse embryonic stem (ES) cells. However, the genes contributing to this stability remain obscure. We previously identified Zscan4 as a specific marker for two-cell embryo and ES cells. Here we show that Zscan4 is involved in telomere maintenance and long-term genomic stability in ES cells. Only 5% of ES cells express Zscan4 at a given time, but nearly all ES cells activate Zscan4 at least once during nine passages. The transient Zscan4-positive state is associated with rapid telomere extension by telomere recombination and upregulation of meiosis-specific homologous recombination genes, which encode proteins that are colocalized with ZSCAN4 on telomeres. Furthermore, Zscan4 knockdown shortens telomeres, increases karyotype abnormalities and spontaneous sister chromatid exchange, and slows down cell proliferation until reaching crisis by passage eight. Together, our data show a unique mode of genome maintenance in ES cells.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Martin, G. R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl Acad. Sci. USA 78, 7634–7638 (1981)
Evans, M. J. & Kaufman, M. H. Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 (1981)
Yoshikawa, T. et al. High-throughput screen for genes predominantly expressed in the ICM of mouse blastocysts by whole mount in situ hybridization. Gene Expr. Patterns 6, 213–224 (2006)
Niwa, H. How is pluripotency determined and maintained? Development 134, 635–646 (2007)
Suda, Y., Suzuki, M., Ikawa, Y. & Aizawa, S. Mouse embryonic stem cells exhibit indefinite proliferative potential. J. Cell. Physiol. 133, 197–201 (1987)
Rebuzzini, P., Neri, T., Zuccotti, M., Redi, C. A. & Garagna, S. Chromosome number variation in three mouse embryonic stem cell lines during culture. Cytotechnology 58, 17–23 (2008)
Longo, L., Bygrave, A., Grosveld, F. G. & Pandolfi, P. P. The chromosome make-up of mouse embryonic stem cells is predictive of somatic and germ cell chimaerism. Transgenic Res. 6, 321–328 (1997)
Nagy, A., Rossant, J., Nagy, R., Abramow-Newerly, W. & Roder, J. C. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl Acad. Sci. USA 90, 8424–8428 (1993)
Cervantes, R. B., Stringer, J. R., Shao, C., Tischfield, J. A. & Stambrook, P. J. Embryonic stem cells and somatic cells differ in mutation frequency and type. Proc. Natl Acad. Sci. USA 99, 3586–3590 (2002)
Blelloch, R. H. et al. Nuclear cloning of embryonal carcinoma cells. Proc. Natl Acad. Sci. USA 101, 13985–13990 (2004)
Brimble, S. N. et al. Karyotypic stability, genotyping, differentiation, feeder-free maintenance, and gene expression sampling in three human embryonic stem cell lines derived prior to August 9, 2001. Stem Cells Dev. 13, 585–597 (2004)
Falco, G. et al. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev. Biol. 307, 539–550 (2007)
Carter, M. G. et al. An in situ hybridization-based screen for heterogeneously expressed genes in mouse ES cells. Gene Expr. Patterns 8, 181–198 (2008)
Storm, M. P. et al. Characterization of the phosphoinositide 3-kinase-dependent transcriptome in murine embryonic stem cells: identification of novel regulators of pluripotency. Stem Cells 27, 764–775 (2009)
Edelstein, L. C. & Collins, T. The SCAN domain family of zinc finger transcription factors. Gene 359, 1–17 (2005)
Feil, R., Wagner, J., Metzger, D. & Chambon, P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem. Biophys. Res. Commun. 237, 752–757 (1997)
Soriano, P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genet. 21, 70–71 (1999)
Doetschman, T. C., Eistetter, H., Katz, M., Schmidt, W. & Kemler, R. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J. Embryol. Exp. Morphol. 87, 27–45 (1985)
Nishiyama, A. et al. Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell 5, 420–433 (2009)
Cawthon, R. M. Telomere measurement by quantitative PCR. Nucleic Acids Res. 30, e47 (2002)
Callicott, R. J. & Womack, J. E. Real-time PCR assay for measurement of mouse telomeres. Comp. Med. 56, 17–22 (2006)
Poon, S. S., Martens, U. M., Ward, R. K. & Lansdorp, P. M. Telomere length measurements using digital fluorescence microscopy. Cytometry 36, 267–278 (1999)
Liu, L. et al. Telomere lengthening early in development. Nature Cell Biol. 9, 1436–1441 (2007)
Bailey, S. M., Goodwin, E. H., Meyne, J. & Cornforth, M. N. CO-FISH reveals inversions associated with isochromosome formation. Mutagenesis 11, 139–144 (1996)
Wang, Y. et al. An increase in telomere sister chromatid exchange in murine embryonic stem cells possessing critically shortened telomeres. Proc. Natl Acad. Sci. USA 102, 10256–10260 (2005)
Dronkert, M. L. et al. Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange. Mol. Cell. Biol. 20, 3147–3156 (2000)
Tateishi, S. et al. Enhanced genomic instability and defective postreplication repair in RAD18 knockout mouse embryonic stem cells. Mol. Cell. Biol. 23, 474–481 (2003)
Keeney, S., Giroux, C. N. & Kleckner, N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997)
Mahadevaiah, S. K. et al. Recombinational DNA double-strand breaks in mice precede synapsis. Nature Genet. 27, 271–276 (2001)
Reinholdt, L. G. & Schimenti, J. C. Mei1 is epistatic to Dmc1 during mouse meiosis. Chromosoma 114, 127–134 (2005)
Revenkova, E. et al. Cohesin SMC1β is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nature Cell Biol. 6, 555–562 (2004)
Chong, L. et al. A human telomeric protein. Science 270, 1663–1667 (1995)
Tanaka, T. S. Transcriptional heterogeneity in mouse embryonic stem cells. Reprod. Fertil. Dev. 21, 67–75 (2009)
Toyooka, Y., Shimosato, D., Murakami, K., Takahashi, K. & Niwa, H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development 135, 909–918 (2008)
Hayashi, K., Lopes, S. M., Tang, F. & Surani, M. A. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 3, 391–401 (2008)
Bailey, S. M., Brenneman, M. A. & Goodwin, E. H. Frequent recombination in telomeric DNA may extend the proliferative life of telomerase-negative cells. Nucleic Acids Res. 32, 3743–3751 (2004)
Niida, H. et al. Telomere maintenance in telomerase-deficient mouse embryonic stem cells: characterization of an amplified telomeric DNA. Mol. Cell. Biol. 20, 4115–4127 (2000)
Bryan, T. M., Englezou, A., Dalla-Pozza, L., Dunham, M. A. & Reddel, R. R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nature Med. 3, 1271–1274 (1997)
Cesare, A. J. & Reddel, R. R. Telomere uncapping and alternative lengthening of telomeres. Mech. Ageing Dev. 129, 99–108 (2008)
De Boeck, G., Forsyth, R. G., Praet, M. & Hogendoorn, P. C. Telomere-associated proteins: cross-talk between telomere maintenance and telomere-lengthening mechanisms. J. Pathol. 217, 327–344 (2009)
Bryan, T. M., Englezou, A., Gupta, J., Bacchetti, S. & Reddel, R. R. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J. 14, 4240–4248 (1995)
Gonzalo, S. et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nature Cell Biol. 8, 416–424 (2006)
Laud, P. R. et al. Elevated telomere-telomere recombination in WRN-deficient, telomere dysfunctional cells promotes escape from senescence and engagement of the ALT pathway. Genes Dev. 19, 2560–2570 (2005)
Ding, H. et al. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell 117, 873–886 (2004)
Aiba, K. et al. Defining developmental potency and cell lineage trajectories by expression profiling of differentiating mouse embryonic stem cells. DNA Res. 16, 73–80 (2009)
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006)
Olson, L. E. et al. Protection from doxorubicin-induced cardiac toxicity in mice with a null allele of carbonyl reductase 1. Cancer Res. 63, 6602–6606 (2003)
Masui, S. et al. An efficient system to establish multiple embryonic stem cell lines carrying an inducible expression unit. Nucleic Acids Res. 33, e43 (2005)
Goodwin, E. & Meyne, J. Strand-specific FISH reveals orientation of chromosome 18 alphoid DNA. Cytogenet. Cell Genet. 63, 126–127 (1993)
Perry, P. & Wolff, S. New Giemsa method for the differential staining of sister chromatids. Nature 251, 156–158 (1974)
Nagy, A., Gertsenstein, M., Vintersten, K. & Behringer, R. Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2003)
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔ T method. Methods 25, 402–408 (2001)
Acknowledgements
We would like to thank T. Hamatani, Y. Nakatake, M. Monti, L. Xin, B. Binder and A. A. Sharov for discussion; Y. Piao, C. Nguyen, D. Eckley, I. Goldberg, D. Dudekula and I. Stanghellini for technical assistance; P. Soriano for providing ROSA26-floxed LacZ cells; and H. Niwa for providing ROSA-tet-inducible system. This work was entirely supported by the Intramural Research Program of the National Institute on Aging, National Institutes of Health.
Author Contributions M.Z., G.F., L.V.S., A.N., M.T., S.-L.L., C.A.S., H.G.H., H.-T.Y., F.E.I. and R.P.W. designed and performed experiments. M.S.H.K. conceived and supervised the project. M.Z. and M.S.H.K. wrote the manuscript. All authors participated in the discussion.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-15 with legends. (PDF 6045 kb)
Rights and permissions
About this article
Cite this article
Zalzman, M., Falco, G., Sharova, L. et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 464, 858–863 (2010). https://doi.org/10.1038/nature08882
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08882
This article is cited by
-
The rules of the totipotency treasure hunt
Nature Cell Biology (2024)
-
The ALT pathway generates telomere fusions that can be detected in the blood of cancer patients
Nature Communications (2024)
-
Low expression of ZSCAN4 predicts unfavorable outcome in urothelial carcinoma of upper urinary tract and urinary bladder
World Journal of Surgical Oncology (2023)
-
ZSCAN4 interacts with PARP1 to promote DNA repair in mouse embryonic stem cells
Cell & Bioscience (2023)
-
Metabolic and cell cycle shift induced by the deletion of Dnm1l attenuates the dissolution of pluripotency in mouse embryonic stem cells
Cellular and Molecular Life Sciences (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.