Abstract
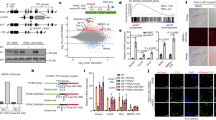
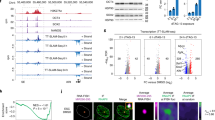
Endogenous retroviruses (ERVs), retrovirus-like elements with long terminal repeats, are widely dispersed in the euchromatic compartment in mammalian cells, comprising ∼10% of the mouse genome1. These parasitic elements are responsible for >10% of spontaneous mutations2. Whereas DNA methylation has an important role in proviral silencing in somatic and germ-lineage cells3,4,5, an additional DNA-methylation-independent pathway also functions in embryonal carcinoma and embryonic stem (ES) cells to inhibit transcription of the exogenous gammaretrovirus murine leukaemia virus (MLV)6,7,8. Notably, a recent genome-wide study revealed that ERVs are also marked by histone H3 lysine 9 trimethylation (H3K9me3) and H4K20me3 in ES cells but not in mouse embryonic fibroblasts9. However, the role that these marks have in proviral silencing remains unexplored. Here we show that the H3K9 methyltransferase ESET (also called SETDB1 or KMT1E) and the Krüppel-associated box (KRAB)-associated protein 1 (KAP1, also called TRIM28)10,11 are required for H3K9me3 and silencing of endogenous and introduced retroviruses specifically in mouse ES cells. Furthermore, whereas ESET enzymatic activity is crucial for HP1 binding and efficient proviral silencing, the H4K20 methyltransferases Suv420h1 and Suv420h2 are dispensable for silencing. Notably, in DNA methyltransferase triple knockout (Dnmt1-/-Dnmt3a-/-Dnmt3b-/-) mouse ES cells, ESET and KAP1 binding and ESET-mediated H3K9me3 are maintained and ERVs are minimally derepressed. We propose that a DNA-methylation-independent pathway involving KAP1 and ESET/ESET-mediated H3K9me3 is required for proviral silencing during the period early in embryogenesis when DNA methylation is dynamically reprogrammed.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
08 April 2010
The accepted date was corrected from 26 January to 16 January 2010 on 8 April 2010.
03 September 2014
Nature 464, 927–931 (2010); doi:10.1038/nature08858 In Fig. 1a of this Letter, it has come to our attention that the lanes for the Dnmt3l+/+ and Dnmt3l−/− testes RNA samples were run on different gels (northern blots) but were not displayed as such. The Dnmt3l−/− sample was originally intended as a positive control for derepression of different retroelements analysed in this paper, such as LINE-1 and IAP.
References
Mouse Genome Sequencing Consortium . Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002)
Maksakova, I. A. et al. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2, e2 (2006)
Walsh, C. P., Chaillet, J. R. & Bestor, T. H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nature Genet. 20, 116–117 (1998)
Yoder, J. A., Walsh, C. P. & Bestor, T. H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13, 335–340 (1997)
Bourc'his, D. & Bestor, T. H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99 (2004)
Teich, N. M., Weiss, R. A., Martin, G. R. & Lowy, D. R. Virus infection of murine teratocarcinoma stem cell lines. Cell 12, 973–982 (1977)
Niwa, O., Yokota, Y., Ishida, H. & Sugahara, T. Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell 32, 1105–1113 (1983)
Pannell, D. et al. Retrovirus vector silencing is de novo methylase independent and marked by a repressive histone code. EMBO J. 19, 5884–5894 (2000)
Mikkelsen, T. S. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007)
Wolf, D. & Goff, S. P. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131, 46–57 (2007)
Wolf, D. & Goff, S. P. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458, 1201–1204 (2009)
Dong, K. B. et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 27, 2691–2701 (2008)
Peters, A. H. et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12, 1577–1589 (2003)
Martens, J. H. et al. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 24, 800–812 (2005)
Tachibana, M. et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 19, 815–826 (2005)
Tsumura, A. et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11, 805–814 (2006)
Maksakova, I. A. & Mager, D. L. Transcriptional regulation of early transposon elements, an active family of mouse long terminal repeat retrotransposons. J. Virol. 79, 13865–13874 (2005)
Schotta, G. et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 18, 1251–1262 (2004)
Kourmouli, N. et al. Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J. Cell Sci. 117, 2491–2501 (2004)
Schultz, D. C., Ayyanathan, K., Negorev, D., Maul, G. G. & Rauscher, F. J. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16, 919–932 (2002)
Ryan, R. F. et al. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol. 19, 4366–4378 (1999)
Schultz, D. C., Friedman, J. R. & Rauscher, F. J. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2α subunit of NuRD. Genes Dev. 15, 428–443 (2001)
Vernet, M. & Cebrian, J. cis-acting elements that mediate the negative regulation of Moloney murine leukemia virus in mouse early embryos. J. Virol. 70, 5630–5633 (1996)
Wolf, D., Hug, K. & Goff, S. P. TRIM28 mediates primer binding site-targeted silencing of Lys1,2 tRNA-utilizing retroviruses in embryonic cells. Proc. Natl Acad. Sci. USA 105, 12521–12526 (2008)
Sarraf, S. A. & Stancheva, I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell 15, 595–605 (2004)
Lachner, M., O’Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001)
Meunier, J., Khelifi, A., Navratil, V. & Duret, L. Homology-dependent methylation in primate repetitive DNA. Proc. Natl Acad. Sci. USA 102, 5471–5476 (2005)
Kim, S. H. et al. Differential DNA methylation reprogramming of various repetitive sequences in mouse preimplantation embryos. Biochem. Biophys. Res. Commun. 324, 58–63 (2004)
Lane, N. et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis 35, 88–93 (2003)
Dodge, J. E., Kang, Y. K., Beppu, H., Lei, H. & Li, E. Histone H3-K9 methyltransferase ESET is essential for early development. Mol. Cell. Biol. 24, 2478–2486 (2004)
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007)
Hawley, R. G., Lieu, F. H., Fong, A. Z. & Hawley, T. S. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1, 136–138 (1994)
Lorincz, M. C. et al. Dynamic analysis of proviral induction and de novo methylation: implications for a histone deacetylase-independent, methylation density-dependent mechanism of transcriptional repression. Mol. Cell. Biol. 20, 842–850 (2000)
Tachibana, M., Matsumura, Y., Fukuda, M., Kimura, H. & Shinkai, Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 27, 2681–2690 (2008)
Kuramochi-Miyagawa, S. et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22, 908–917 (2008)
Acknowledgements
We thank M. Okano for Dnmt1-/-Dnmt3a-/-Dnmt3b-/- ES cells; T. Jenuwein and P. Singh for Suv39h1-/-Suv39h2-/- ES cells; T. Jenuwein and G. Schotta for Suv420h1-/-Suv420h2-/- ES cells; K. Hata for Dnmt3l knockout mice testes; M. Hijikata for the large-T antigen expression vector; S. Kuramochi-Miyagawa for IAP and LINE-1 probes; S. Smale for HP1α antiserum; and L. Gaudreau for H2AZ antiserum. We are also grateful to P. Goyal and members of the Shinkai laboratory for technical support and D. Mager for critically reading the manuscript. This work was supported in part by the Genome Network Project from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, Grants-in-aid from MEXT to Y.S. and CIHR grants 77805 and 92090 to M.C.L. M.C.L. is a Scholar of the Michael Smith Foundation for Health Research.
Author Contributions T.M. and D.L. are equally contributing first authors; M.C.L. and Y.S. are equally contributing senior authors. Y.S., M.C.L., D.L. and T.M. planned studies and interpreted the data. T.M. validated the Eset CKO lines and performed most of the ERV-related studies. D.L. performed the siRNA and exogenous retrovirus-related experiments. H. Miyashita generated Eset CKO ES cells and mice. I.A.M. conducted the MusD sequencing analysis. H. Miyachi injected Eset CKO ES cells into blastocysts to make chimaeric mice. H.K. provided antibodies against methylated histones. M.T. cloned mouse Eset cDNA and provided his expertise in most of the studies conducted in this work. Y.S., M.C.L., D.L. and T.M. wrote the paper.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion, Supplementary Figures S1-S17 with Legends and Supplementary References. (PDF 16352 kb)
Supplementary Table 1
This Supplementary Table file contains a Primer list. (XLS 34 kb)
Rights and permissions
About this article
Cite this article
Matsui, T., Leung, D., Miyashita, H. et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931 (2010). https://doi.org/10.1038/nature08858
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08858
This article is cited by
-
Cell-type differential targeting of SETDB1 prevents aberrant CTCF binding, chromatin looping, and cis-regulatory interactions
Nature Communications (2024)
-
Keep quiet: the HUSH complex in transcriptional silencing and disease
Nature Structural & Molecular Biology (2024)
-
Auto-suppression of Tet dioxygenases protects the mouse oocyte genome from oxidative demethylation
Nature Structural & Molecular Biology (2024)
-
The functions of SET domain bifurcated histone lysine methyltransferase 1 (SETDB1) in biological process and disease
Epigenetics & Chromatin (2023)
-
Differential effect of histone H3.3 depletion on retroviral repression in embryonic stem cells
Clinical Epigenetics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.