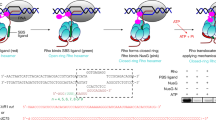
Abstract
Rho is the essential RNA helicase that sets the borders between transcription units and adjusts transcriptional yield to translational needs in bacteria1,2,3. Although Rho was the first termination factor to be discovered4, the actual mechanism by which it reaches and disrupts the elongation complex (EC) is unknown. Here we show that the termination-committed Rho molecule associates with RNA polymerase (RNAP) throughout the transcription cycle; that is, it does not require the nascent transcript for initial binding. Moreover, the formation of the RNAP–Rho complex is crucial for termination. We show further that Rho-dependent termination is a two-step process that involves rapid EC inactivation (trap) and a relatively slow dissociation. Inactivation is the critical rate-limiting step that establishes the position of the termination site. The trap mechanism depends on the allosterically induced rearrangement of the RNAP catalytic centre by means of the evolutionarily conserved mobile trigger-loop domain, which is also required for EC dissociation. The key structural and functional similarities, which we found between Rho-dependent and intrinsic (Rho-independent) termination pathways, argue that the allosteric mechanism of termination is general and likely to be preserved for all cellular RNAPs throughout evolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Adhya, S. & Gottesman, M. Control of transcription termination. Annu. Rev. Biochem. 47, 967–996 (1978)
Richardson, J. P. Preventing the synthesis of unused transcripts by Rho factor. Cell 64, 1047–1049 (1991)
Cardinale, C. J. et al. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli . Science 320, 935–938 (2008)
Roberts, J. W. Termination factor for RNA synthesis. Nature 224, 1168–1174 (1969)
Geiselmann, J., Yager, T. D., Gill, S. C., Calmettes, P. & von Hippel, P. H. Physical properties of the Escherichia coli transcription termination factor Rho. 1. Association states and geometry of the Rho hexamer. Biochemistry 31, 111–121 (1992)
Browne, R. J., Barr, E. W. & Stitt, B. L. Catalytic cooperativity among subunits of Escherichia coli transcription termination factor Rho. Kinetics and substrate structural requirements. J. Biol. Chem. 280, 13292–13299 (2005)
Skordalakes, E. & Berger, J. M. Structural insights into RNA-dependent ring closure and ATPase activation by the Rho termination factor. Cell 127, 553–564 (2006)
Adelman, J. L. et al. Mechanochemistry of transcription termination factor Rho. Mol. Cell 22, 611–621 (2006)
Banerjee, S., Chalissery, J., Bandey, I. & Sen, R. Rho-dependent transcription termination: more questions than answers. J. Microbiol. 44, 11–22 (2006)
Platt, T. Rho and RNA: models for recognition and response. Mol. Microbiol. 11, 983–990 (1994)
Kim, D. E. & Patel, S. S. The kinetic pathway of RNA binding to the Escherichia coli transcription termination factor Rho. J. Biol. Chem. 276, 13902–13910 (2001)
Roberts, J. W., Shankar, S. & Filter, J. J. RNA polymerase elongation factors. Annu. Rev. Microbiol. 62, 211–233 (2008)
Jin, D. J., Burgess, R. R., Richardson, J. P. & Gross, C. A. Termination efficiency at Rho-dependent terminators depends on kinetic coupling between RNA polymerase and Rho. Proc. Natl Acad. Sci. USA 89, 1453–1457 (1992)
Zhu, A. Q. & von Hippel, P. H. Rho-dependent termination within the trp t′ terminator. II. Effects of kinetic competition and rho processivity. Biochemistry 37, 11215–11222 (1998)
Guerin, M., Robichon, N., Geiselmann, J. & Rahmouni, A. R. A simple polypyrimidine repeat acts as an artificial rho-dependent terminator in vivo and in vitro . Nucleic Acids Res. 26, 4895–4900 (1998)
Chalissery, J., Banerjee, S., Bandey, I. & Sen, R. Transcription termination defective mutants of Rho: role of different functions of Rho in releasing RNA from the elongation complex. J. Mol. Biol. 371, 855–872 (2007)
Bar-Nahum, G., Epshtein, V., Ruckenstein, A. E., Mustaev, A. & Nudler, E. A ratchet mechanism of transcription elongation and its control. Cell 120, 183–193 (2005)
Vassylyev, D. G. et al. Structural basis for transcription inhibition by tagetitoxin. Nature Struct. Mol. Biol. 12, 1086–1093 (2005)
Mentesana, P. E., Chin-Bow, S. T., Sousa, R. & McAllister, W. T. Characterization of halted T7 RNA polymerase elongation complexes reveals multiple factors that contribute to stability. J. Mol. Biol. 302, 1049–1062 (2000)
Kettenberger, H., Armache, K. J. & Cramer, P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell 16, 955–965 (2004)
Wang, D., Bushnell, D. A., Westover, K. D., Kaplan, C. D. & Kornberg, R. D. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell 127, 941–954 (2006)
Vassylyev, D. G., Vassylyeva, M. N., Perederina, A., Tahirov, T. H. & Artsimovitch, I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature 448, 157–162 (2007)
Toulokhonov, I., Zhang, J., Palangat, M. & Landick, R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol. Cell 27, 406–419 (2007)
Brennan, C. A., Dombroski, A. J. & Platt, T. Transcription termination factor rho is an RNA-DNA helicase. Cell 48, 945–952 (1987)
Schwartz, A., Margeat, E., Rahmouni, A. R. & Boudvillain, M. Transcription termination factor rho can displace streptavidin from biotinylated RNA. J. Biol. Chem. 282, 1469–1476 (2007)
Gusarov, I. & Nudler, E. The mechanism of intrinsic transcription termination. Mol. Cell 3, 495–504 (1999)
Komissarova, N., Becker, J., Solter, S., Kireeva, M. & Kashlev, M. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol. Cell 10, 1151–1162 (2002)
Epshtein, V., Cardinale, C., Ruckenstein, A., Borukhov, S. & Nudler, E. An allosteric path to transcription termination. Mol. Cell 28, 991–1001 (2007)
Westover, K. D., Bushnell, D. A. & Kornberg, R. D. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science 303, 1014–1016 (2004)
Naryshkina, T., Kuznedelov, K. & Severinov, K. The role of the largest RNA polymerase subunit lid element in preventing the formation of extended RNA-DNA hybrid. J. Mol. Biol. 361, 634–643 (2006)
Nudler, E., Gusarov, I. & Bar-Nahum, G. Methods of walking with the RNA polymerase. Methods Enzymol. 371, 160–169 (2003)
Wright, D. J., King, K. & Modrich, P. The negative charge of Glu-111 is required to activate the cleavage center of EcoRI endonuclease. J. Biol. Chem. 264, 11816–11821 (1989)
Borukhov, S. & Goldfarb, A. Recombinant Escherichia coli RNA polymerase: purification of individually overexpressed subunits and in vitro assembly. Protein Expr. Purif. 4, 503–511 (1993)
Nudler, E., Gusarov, I., Avetissova, E., Kozlov, M. & Goldfarb, A. Spatial organization of transcription elongation complex in Escherichia coli . Science 281, 424–428 (1998)
Acknowledgements
We thank I. Artsimovitch, S. Borukhov, K. Severinov, B. Stitt, D. Vassylyev and R. Weisberg for materials and discussions. This work was supported by a grant from the National Institutes of Health (R01GM58750, to E.N.).
Author Contributions V.E., D.D. and J.W. conducted the experimental work, discussed the results and commented on the manuscript. E.N. designed the study and wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures, 1-11 with Legends and Supplementary Notes and Supplementary Methods. (PDF 1700 kb)
Rights and permissions
About this article
Cite this article
Epshtein, V., Dutta, D., Wade, J. et al. An allosteric mechanism of Rho-dependent transcription termination. Nature 463, 245–249 (2010). https://doi.org/10.1038/nature08669
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature08669
This article is cited by
-
Pervasive Transcription-coupled DNA repair in E. coli
Nature Communications (2022)
-
Rho-dependent transcription termination proceeds via three routes
Nature Communications (2022)
-
Cryo-EM structure of transcription termination factor Rho from Mycobacterium tuberculosis reveals bicyclomycin resistance mechanism
Communications Biology (2022)
-
Efficient production of long double-stranded RNAs applicable to agricultural pest control by Corynebacterium glutamicum equipped with coliphage T7-expression system
Applied Microbiology and Biotechnology (2021)
-
RhoTermPredict: an algorithm for predicting Rho-dependent transcription terminators based on Escherichia coli, Bacillus subtilis and Salmonella enterica databases
BMC Bioinformatics (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.