Abstract
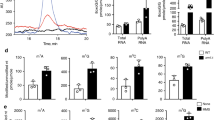
Translational fidelity, essential for protein and cell function, requires accurate transfer RNA (tRNA) aminoacylation. Purified aminoacyl-tRNA synthetases exhibit a fidelity of one error per 10,000 to 100,000 couplings1,2. The accuracy of tRNA aminoacylation in vivo is uncertain, however, and might be considerably lower3,4,5,6. Here we show that in mammalian cells, approximately 1% of methionine (Met) residues used in protein synthesis are aminoacylated to non-methionyl-tRNAs. Remarkably, Met-misacylation increases up to tenfold upon exposing cells to live or non-infectious viruses, toll-like receptor ligands or chemically induced oxidative stress. Met is misacylated to specific non-methionyl-tRNA families, and these Met-misacylated tRNAs are used in translation. Met-misacylation is blocked by an inhibitor of cellular oxidases, implicating reactive oxygen species (ROS) as the misacylation trigger. Among six amino acids tested, tRNA misacylation occurs exclusively with Met. As Met residues are known to protect proteins against ROS-mediated damage7, we propose that Met-misacylation functions adaptively to increase Met incorporation into proteins to protect cells against oxidative stress. In demonstrating an unexpected conditional aspect of decoding mRNA, our findings illustrate the importance of considering alternative iterations of the genetic code.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ibba, M. & Soll, D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 (2000)
Cochella, L. & Green, R. Fidelity in protein synthesis. Curr. Biol. 15, R536–R540 (2005)
Lee, J. W. et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 443, 50–55 (2006)
Miranda, I., Silva, R. & Santos, M. A. Evolution of the genetic code in yeasts. Yeast 23, 203–213 (2006)
Silva, R. M. et al. Critical roles for a genetic code alteration in the evolution of the genus Candida . EMBO J. 26, 4555–4565 (2007)
Ahel, I., Korencic, D., Ibba, M. & Soll, D. Trans-editing of mischarged tRNAs. Proc. Natl Acad. Sci. USA 100, 15422–15427 (2003)
Luo, S. & Levine, R. L. Methionine in proteins defends against oxidative stress. FASEB J. 23, 464–472 (2009)
Dittmar, K. A., Goodenbour, J. M. & Pan, T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2, e221 (2006)
Ussery, M. A., Tanaka, W. K. & Hardesty, B. Subcellular distribution of aminoacyl-tRNA synthetases in various eukaryotic cells. Eur. J. Biochem. 72, 491–500 (1977)
Varshavsky, A. Regulated protein degradation. Trends Biochem. Sci. 30, 283–286 (2005)
Ibba, M. & Soll, D. Quality control mechanisms during translation. Science 286, 1893–1897 (1999)
Nishimura, M. & Naito, S. Tissue-specific mRNA expression profiles of human toll-like receptors and related genes. Biol. Pharm. Bull. 28, 886–892 (2005)
Malhotra, J. D. et al. Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proc. Natl Acad. Sci. USA 105, 18525–18530 (2008)
Murray, J. I. et al. Diverse and specific gene expression responses to stresses in cultured human cells. Mol. Biol. Cell 15, 2361–2374 (2004)
Levine, R. L., Mosoni, L., Berlett, B. S. & Stadtman, E. R. Methionine residues as endogenous antioxidants in proteins. Proc. Natl Acad. Sci. USA 93, 15036–15040 (1996)
Oien, D. B. & Moskovitz, J. Substrates of the methionine sulfoxide reductase system and their physiological relevance. Curr. Top. Dev. Biol. 80, 93–133 (2008)
Chernyak, B. V. e. t. a. l. Production of reactive oxygen species in mitochondria of HeLa cells under oxidative stress. Biochim. Biophys. Acta Bioenergetics 1757, 525–534 (2006)
Savina, A. et al. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell 126, 205–218 (2006)
Bender, A., Hajieva, P. & Moosmann, B. Adaptive antioxidant methionine accumulation in respiratory chain complexes explains the use of a deviant genetic code in mitochondria. Proc. Natl Acad. Sci. USA 105, 16496–16501 (2008)
Finkelstein, J. D. Metabolic regulatory properties of S-adenosylmethionine and S-adenosylhomocysteine. Clin. Chem. Lab. Med. 45, 1694–1699 (2007)
Ruan, B. et al. Quality control despite mistranslation caused by an ambiguous genetic code. Proc. Natl Acad. Sci. USA 105, 16502–16507 (2008)
Santos, M. A., Cheesman, C., Costa, V., Moradas-Ferreira, P. & Tuite, M. F. Selective advantages created by codon ambiguity allowed for the evolution of an alternative genetic code in Candida spp. Mol. Microbiol. 31, 937–947 (1999)
Hoffmann, G. W. On the origin of the genetic code and the stability of the translation apparatus. J. Mol. Biol. 86, 349–362 (1974)
Freist, W., Sternbach, H., Pardowitz, I. & Cramer, F. Accuracy of protein biosynthesis: quasi-species nature of proteins and possibility of error catastrophes. J. Theor. Biol. 193, 19–38 (1998)
Acknowledgements
The authors are grateful to D. Klinman for his gift of CpG oligonucleotides and advice, A. Schilling for supervision and advice on mass spectrometry experiments, and C. Nicchitta, T. Pierson, P. Cluzel, R. Levine, A. Iwasaki and S. Amigorena for insight and advice. This work was supported by the Division of Intramural Research, the National Institute of Allergy and Infectious Diseases, and by National Institutes of Health extramural pilot projects.
Author Contributions N.N., J.M.G., A.D., K.A.D., R.B.J., J.R.S., D.B., E.M.E, M.R.R., J.S.B., A.E., B.D., S.D., H.D.H., P.B. and T.P. designed and performed experiments, and analysed data. J.S.G. generated genetic constructs. J.R.B. and J.W.Y designed experiments and analysed data. J.W.Y. and T.P. conceived the project and wrote the paper. J.W.Y. and T.P. contributed equally to this paper.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Figures
This file contains Supplementary Figures S1-S19 with Legends. (PDF 1207 kb)
Supplementary Information
This file contains Supplementary Methods and Supplementary References and was added online on February 2, 2010. (PDF 196 kb)
Rights and permissions
About this article
Cite this article
Netzer, N., Goodenbour, J., David, A. et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature 462, 522–526 (2009). https://doi.org/10.1038/nature08576
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature08576
This article is cited by
-
Bias at the third nucleotide of codon pairs in virus and host genomes
Scientific Reports (2022)
-
Tryptophan depletion results in tryptophan-to-phenylalanine substitutants
Nature (2022)
-
A guide to antigen processing and presentation
Nature Reviews Immunology (2022)
-
Challenges in Analysis of Hydrophilic Metabolites Using Chromatography Coupled with Mass Spectrometry
Journal of Analysis and Testing (2020)
-
The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis
Nature Reviews Molecular Cell Biology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.