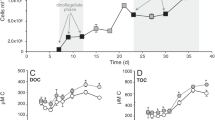
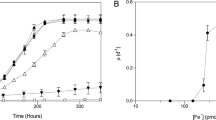
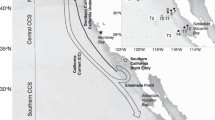
Abstract
Coastal waters support ∼90 per cent of global fisheries and are therefore an important food reserve for our planet1. Eutrophication of these waters, due to human activity, leads to severe oxygen depletion and the episodic occurrence of hydrogen sulphide—toxic to multi-cellular life—with disastrous consequences for coastal ecosytems2,3,4,5. Here we show that an area of ∼7,000 km2 of African shelf, covered by sulphidic water, was detoxified by blooming bacteria that oxidized the biologically harmful sulphide to environmentally harmless colloidal sulphur and sulphate. Combined chemical analyses, stoichiometric modelling, isotopic incubations, comparative 16S ribosomal RNA, functional gene sequence analyses and fluorescence in situ hybridization indicate that the detoxification proceeded by chemolithotrophic oxidation of sulphide with nitrate and was mainly catalysed by two discrete populations of γ- and ε-proteobacteria. Chemolithotrophic bacteria, accounting for ∼20 per cent of the bacterioplankton in sulphidic waters, created a buffer zone between the toxic sulphidic subsurface waters and the oxic surface waters, where fish and other nekton live. This is the first time that large-scale detoxification of sulphidic waters by chemolithotrophs has been observed in an open-ocean system. The data suggest that sulphide can be completely consumed by bacteria in the subsurface waters and, thus, can be overlooked by remote sensing or monitoring of shallow coastal waters. Consequently, sulphidic bottom waters on continental shelves may be more common than previously believed, and could therefore have an important but as yet neglected effect on benthic communities.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Pauly, D. et al. Towards sustainability in world fisheries. Nature 418, 689–695 (2002)
Diaz, R. J. & Rosenberg, R. Marine benthic hypoxia: a review of its ecological effects and the behavioral responses of benthic macrofauna. Oceanogr. Mar. Biol. Annu. Rev. 33, 245–303 (1995)
Naqvi, S. W. A. et al. Increased marine production of N2O due to intensifying anoxia on the Indian continental shelf. Nature 408, 346–349 (2000)
Rabalais, N. N., Turner, R. E. & Scavia, D. Beyond science into policy: Gulf of Mexico hypoxia and the Mississippi River. Bioscience 52, 129–142 (2002)
Malakoff, D. Death by suffocation in the Gulf of Mexico. Science 281, 190–192 (1998)
Carr, M.-E. Estimation of potential productivity in eastern boundary currents using remote sensing. Deep-Sea Res. II 49, 58–80 (2002)
Copenhagen, W. J. The periodic mortality of fish in the Walvis Bay region. Investl Rep. Div. Sea Fish S. Afr. 14, 1–35 (1953)
Hart, T. J. & Currie, R. I. The Benguela current. Discovery Rep. 31, 123–297 (1960)
Emeis, K. C. et al. Shallow gas in shelf sediments of the Namibian coastal upwelling ecosystem. Continent. Shelf Res. 24, 627–642 (2004)
Weeks, S. J., Currie, B. & Bakun, A. Satellite imaging: Massive emissions of toxic gas in the Atlantic. Nature 415, 493–494 (2002)
Bruchert, V. et al. in Past and Present Water Column Anoxia (ed. Neretin, L. N.) 161–193 (NATO Science Series 40, Springer, 2006)
Berg, P., Risgaard-Petersen, N. & Rysgaard, S. Interpretation of measured concentration profiles in sediment pore water. Limnol. Oceanogr. 43, 1500–1510 (1998)
Kuypers, M. M. M. et al. Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc. Natl Acad. Sci. USA 102, 6478–6483 (2005)
Cardoso, R. B. et al. Sulfide oxidation under chemolithoautotrophic denitrifying conditions. Biotechnol. Bioeng. 95, 1148–1157 (2006)
Peek, A. S., Feldman, R. A., Lutz, R. A. & Vrijenhoek, R. C. Cospeciation of chemoautotrophic bacteria and deep sea clams. Proc. Natl Acad. Sci. USA 95, 9962–9966 (1998)
Newton, I. L. G. et al. The Calyptogena magnifica chemoautotrophic symbiont genome. Science 315, 998–1000 (2007)
Madrid, V. M., Taylor, G. T., Scranton, M. I. & Chistoserdov, A. Y. Phylogenetic diversity of bacterial and archaeal communities in the anoxic zone of the Cariaco basin. Appl. Environ. Microbiol. 67, 1663–1674 (2001)
Sunamura, M., Higashi, Y., Miyako, C., Ishibashi, J. & Maruyama, A. Two bacteria phylotypes are predominant in the Suiyo seamount hydrothermal plume. Appl. Environ. Microbiol. 70, 1190–1198 (2004)
Vetter, R. D. Elemental sulfur in the gills of three species of clams containing chemoautotrophic symbiotic bacteria: a possible inorganic energy storage compound. Mar. Biol. 88, 33–42 (1985)
Campbell, B. J. et al. The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat. Rev. Microbiol. 4, 458–468 (2006)
Wirsen, C. O. et al. Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp that produces filamentous sulphur. Appl. Environ. Microbiol. 68, 316–325 (2002)
Sievert, S. M. et al. Genome of the Epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans . Appl. Environ. Microbiol. 74, 1145–1156 (2008)
Gevertz, D. et al. Isolation and characterization of strains CVO and FWKO B, two novel nitrate-reducing, sulfide-oxidizing bacteria isolated from oil field brine. Appl. Environ. Microbiol. 66, 2491–2501 (2000)
Beman, J. M., Arrigo, K. R. & Matson, P. A. Agricultural runoff fuels large phytoplankton blooms in vulnerable areas of the ocean. Nature 434, 211–214 (2005)
Grantham, B. A. et al. Upwelling-driven nearshore hypoxia signals ecosystem and oceanographic changes in the northeast Pacific. Nature 429, 749–754 (2004)
Sarmiento, J. L. et al. Simulated response of the ocean carbon cycle to anthropogenic climate warming. Nature 393, 245–249 (1998)
Paerl, H. W. & Steppe, T. F. Scaling up: the next challenge in environmental microbiology. Environ. Microbiol. 5, 1025–1038 (2003)
Ohde, T. et al. Identification and investigation of sulphur plumes along the Namibian coast using the MERIS sensor. Continent. Shelf Res. 27, 744–756 (2007)
Ludwig, W. et al. ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371 (2004)
Cline, J. D. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14, 454–458 (1969)
Hansen, J. W., Thamdrup, B. & Jorgensen, B. B. Anoxic incubation of sediment in gas-tight plastic bags: a method for biogeochemical process studies. Mar. Ecol. Prog. Ser. 208, 273–282 (2000)
Thamdrup, B. & Dalsgaard, T. Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl. Environ. Microbiol. 68, 1312–1318 (2002)
Kallmeyer, J., Ferdelman, T. G., Weber, A., Fossing, H. & Jørgensen, B. B. A cold chromium distillation procedure for radiolabeled sulfide applied to sulfate reduction measurements. Limnol. Oceanogr. Methods 2, 171–180 (2004)
Blazejak, A. et al. Phylogeny of 16S rRNA, ribulose 1,5-bisphosphate carboxylase/oxygenase, and adenosine 5′-phosphosulfate reductase genes from gamma- and alphaproteobacterial symbionts in gutless marine worms (Oligochaeta) from Bermuda and the Bahamas. Appl. Environ. Microbiol. 72, 5527–5536 (2006)
Kirchman, D. L. et al. Structure of bacterial communities in aquatic systems as revealed by filter PCR. Aquat. Microb. Ecol. 26, 13–22 (2001)
Zhou, J., Bruns, M. A. & Tiedje, J. M. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62, 316–322 (1996)
Pernthaler, A., Pernthaler, J. & Amann, R. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68, 3094–3101 (2002)
Snaidr, J. et al. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63, 2884–2896 (1997)
Gloeckner, F. O. et al. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst. Appl. Microbiol. 19, 403–406 (1996)
Fennel, K. & Boss, E. Subsurface maxima of phytoplankton and chlorophyll: Steady-state solutions from a simple model. Limnol. Oceanogr. 48, 1521–1534 (2003)
Acknowledgements
We thank B. Barker Jørgensen, F. Inagaki, C. Hubert, T. Ferdelman and G. Collins for discussions; the Namibian authorities for access to their national waters; the crew of RV Alexander von Humboldt for assistance onboard; S. Krüger for operating the pump-CTD and oceanographic equipment; T. Heene for assistance with the collection of oceanographic data and generating oceanographic plots; and G. Klockgether, J. Sawicka, J. Wulf, S. Lenk, D. Franzke and K. Nkandi for assistance with the analysis. The investigations were supported by the MPG, the BMBF programme Geotechnologien and the project NAMIBGAS, the DFG, BENEFIT and the Namibian Ministry of Fisheries and Natural Resources.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Table 1, a Supplementary Discussion, Supplementary Figures 1- 4 with Legends and Supplementary References. (PDF 318 kb)
Rights and permissions
About this article
Cite this article
Lavik, G., Stührmann, T., Brüchert, V. et al. Detoxification of sulphidic African shelf waters by blooming chemolithotrophs. Nature 457, 581–584 (2009). https://doi.org/10.1038/nature07588
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07588
This article is cited by
-
Magnetotactic bacteria and magnetofossils: ecology, evolution and environmental implications
npj Biofilms and Microbiomes (2022)
-
Changes in the membrane lipid composition of a Sulfurimonas species depend on the electron acceptor used for sulfur oxidation
ISME Communications (2022)
-
Microbial production of toluene in oxygen minimum zone waters in the Humboldt Current System off Chile
Scientific Reports (2022)
-
Niche differentiation of sulfur-oxidizing bacteria (SUP05) in submarine hydrothermal plumes
The ISME Journal (2022)
-
Carbon assimilating fungi from surface ocean to subseafloor revealed by coupled phylogenetic and stable isotope analysis
The ISME Journal (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.